单个氨基酸的有害变化对推断 AAV2 和 AAV2-13 外壳适应性的预测能力
IF 4.6
2区 医学
Q2 MEDICINE, RESEARCH & EXPERIMENTAL
Molecular Therapy-Methods & Clinical Development
Pub Date : 2024-08-20
DOI:10.1016/j.omtm.2024.101327
引用次数: 0
摘要
腺相关病毒(AAV)是应用最广泛的基因转移载体。噬菌体工程的一个主要局限是对 AAV 噬菌体稳定性的多个氨基酸变异的后果了解不全面,导致噬菌体无法存活的频率很高。在这种情况下,对天然 AAV 变体的研究可以为了解对突变具有更大耐受性的囊膜区域提供宝贵的见解。在这里,对 AAV2 变体的特征描述和对两个公共噬菌体文库的分析凸显了与有害突变相关的共同特征,表明突变对噬菌体活力的影响严格依赖于它们在噬菌体结构中的三维位置。我们开发了一种新的预测方法来推断含有多个氨基酸变异的AAV2变体的适应性,灵敏度为98%,准确度为98%,特异性为95%。这种新方法可以简化开发富含有活力噬菌体的 AAV 载体文库的过程,从而加快从工程噬菌体中鉴定候选治疗药物的速度。本文章由计算机程序翻译,如有差异,请以英文原文为准。
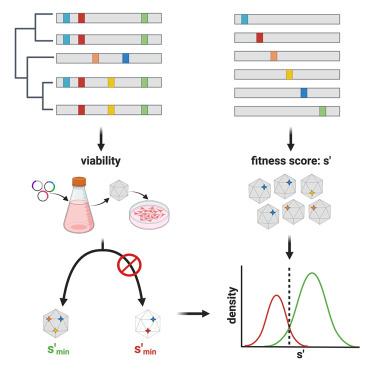
Predictive power of deleterious single amino acid changes to infer on AAV2 and AAV2-13 capsids fitness
Adeno-associated virus (AAV) is the most widely used vector for gene transfer. A major limitation of capsid engineering is the incomplete understanding of the consequences of multiple amino acid variations on AAV capsid stability resulting in high frequency of non-viable capsids. In this context, the study of natural AAV variants can provide valuable insights into capsid regions that exhibit greater tolerance to mutations. Here, the characterization of AAV2 variants and the analysis of two public capsid libraries highlighted common features associated with deleterious mutations, suggesting that the impact of mutations on capsid viability is strictly dependent on their 3D location within the capsid structure. We developed a novel prediction method to infer the fitness of AAV2 variants containing multiple amino acid variations with 98% sensitivity, 98% accuracy, and 95% specificity. This novel approach might streamline the development of AAV vector libraries enriched in viable capsids, thus accelerating the identification of therapeutic candidates among engineered capsids.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Therapy-Methods & Clinical Development
Biochemistry, Genetics and Molecular Biology-Molecular Biology
CiteScore
9.90
自引率
4.30%
发文量
163
审稿时长
12 weeks
期刊介绍:
The aim of Molecular Therapy—Methods & Clinical Development is to build upon the success of Molecular Therapy in publishing important peer-reviewed methods and procedures, as well as translational advances in the broad array of fields under the molecular therapy umbrella.
Topics of particular interest within the journal''s scope include:
Gene vector engineering and production,
Methods for targeted genome editing and engineering,
Methods and technology development for cell reprogramming and directed differentiation of pluripotent cells,
Methods for gene and cell vector delivery,
Development of biomaterials and nanoparticles for applications in gene and cell therapy and regenerative medicine,
Analysis of gene and cell vector biodistribution and tracking,
Pharmacology/toxicology studies of new and next-generation vectors,
Methods for cell isolation, engineering, culture, expansion, and transplantation,
Cell processing, storage, and banking for therapeutic application,
Preclinical and QC/QA assay development,
Translational and clinical scale-up and Good Manufacturing procedures and process development,
Clinical protocol development,
Computational and bioinformatic methods for analysis, modeling, or visualization of biological data,
Negotiating the regulatory approval process and obtaining such approval for clinical trials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


