用于精确表达的慢病毒载体治疗 X 连锁淋巴细胞增生症
IF 4.6
2区 医学
Q2 MEDICINE, RESEARCH & EXPERIMENTAL
Molecular Therapy-Methods & Clinical Development
Pub Date : 2024-08-20
DOI:10.1016/j.omtm.2024.101323
引用次数: 0
摘要
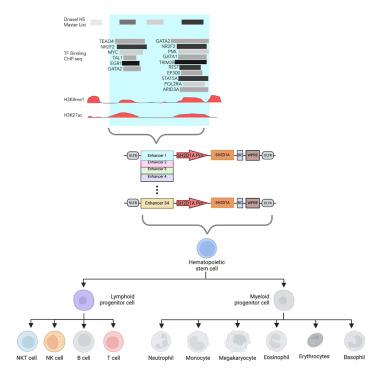
X连锁淋巴细胞增生症(XLP1)是由影响SLAM相关蛋白(SAP)的基因突变引起的。设计用于在 T、NK 和 NKT 细胞中以治疗水平表达 SAP 的调控慢病毒载体(LV)XLP-SMART LV 对于有效的基因治疗至关重要。我们通过实验确定了该基因的 34 个基因组调控元件,并设计了 XLP-SMART LV 来模拟 SAP 的系谱和阶段特异性调控。我们筛选了它们在 T 细胞、NK 细胞和 NKT 细胞中的靶上增强子活性,以及在 B 细胞和骨髓细胞群中的脱靶增强子活性。三个增强子元件组合在一起可使SAP启动子在靶上群体中的表达量增加4倍。使用XLP-SMART LV进行的NSG-Tg(Hu-IL15)异种移植研究表明,与对照组EFS-LV相比,SAP在靶细胞中的表达量增加了7倍,而且没有脱靶表达。XLP-SMART LV 在外周血、骨髓、脾脏和胸腺组织中表现出阶段特异性的 T 细胞和 NK 细胞表达(模拟 SAP 的表达模式)。用XLP-SMART LVs转导XLP1患者的CD8+ T细胞或BM CD34+细胞,可分别将刺激诱导的细胞死亡和NK细胞毒性恢复到野生型水平。这些数据表明,通过基因疗法创建一种系和阶段特异性 LV 来恢复 XLP1 表型是可行的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Lentiviral vectors for precise expression to treat X-linked lymphoproliferative disease
X-linked lymphoproliferative disease (XLP1) results from gene mutations affecting the SLAM-associated protein (SAP). A regulated lentiviral vector (LV), XLP-SMART LV, designed to express SAP at therapeutic levels in T, NK, and NKT cells, is crucial for effective gene therapy. We experimentally identified 34 genomic regulatory elements of the gene and designed XLP-SMART LVs to emulate the lineage and stage-specific control of SAP. We screened them for their on-target enhancer activity in T, NK, and NKT cells and their off-target enhancer activity in B cell and myeloid populations. In combination, three enhancer elements increased SAP promoter expression up to 4-fold in on-target populations . NSG-Tg(Hu-IL15) xenograft studies with XLP-SMART LVs demonstrated up to 7-fold greater expression in on-target cells over a control EFS-LV, with no off-target expression. The XLP-SMART LVs exhibited stage-specific T and NK cell expression in peripheral blood, bone marrow, spleen, and thymic tissues (mimicking expression patterns of SAP). Transduction of XLP1 patient CD8+ T cells or BM CD34+ cells with XLP-SMART LVs restored restimulation-induced cell death and NK cytotoxicity to wild-type levels, respectively. These data demonstrate that it is feasible to create a lineage and stage-specific LV to restore the XLP1 phenotype by gene therapy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Therapy-Methods & Clinical Development
Biochemistry, Genetics and Molecular Biology-Molecular Biology
CiteScore
9.90
自引率
4.30%
发文量
163
审稿时长
12 weeks
期刊介绍:
The aim of Molecular Therapy—Methods & Clinical Development is to build upon the success of Molecular Therapy in publishing important peer-reviewed methods and procedures, as well as translational advances in the broad array of fields under the molecular therapy umbrella.
Topics of particular interest within the journal''s scope include:
Gene vector engineering and production,
Methods for targeted genome editing and engineering,
Methods and technology development for cell reprogramming and directed differentiation of pluripotent cells,
Methods for gene and cell vector delivery,
Development of biomaterials and nanoparticles for applications in gene and cell therapy and regenerative medicine,
Analysis of gene and cell vector biodistribution and tracking,
Pharmacology/toxicology studies of new and next-generation vectors,
Methods for cell isolation, engineering, culture, expansion, and transplantation,
Cell processing, storage, and banking for therapeutic application,
Preclinical and QC/QA assay development,
Translational and clinical scale-up and Good Manufacturing procedures and process development,
Clinical protocol development,
Computational and bioinformatic methods for analysis, modeling, or visualization of biological data,
Negotiating the regulatory approval process and obtaining such approval for clinical trials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


