缩小开源和商业大型语言模型在医学证据摘要方面的差距
IF 12.4
1区 医学
Q1 HEALTH CARE SCIENCES & SERVICES
引用次数: 0
摘要
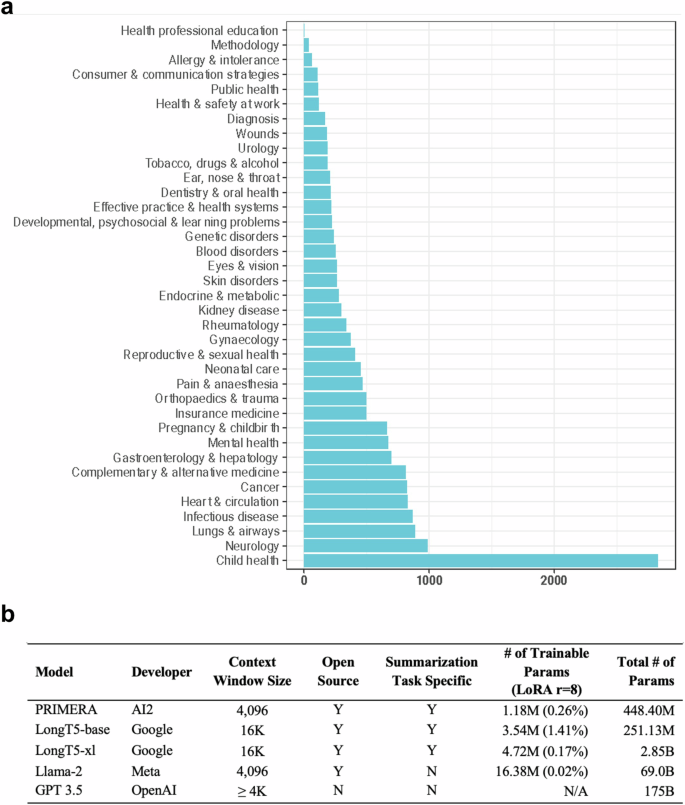
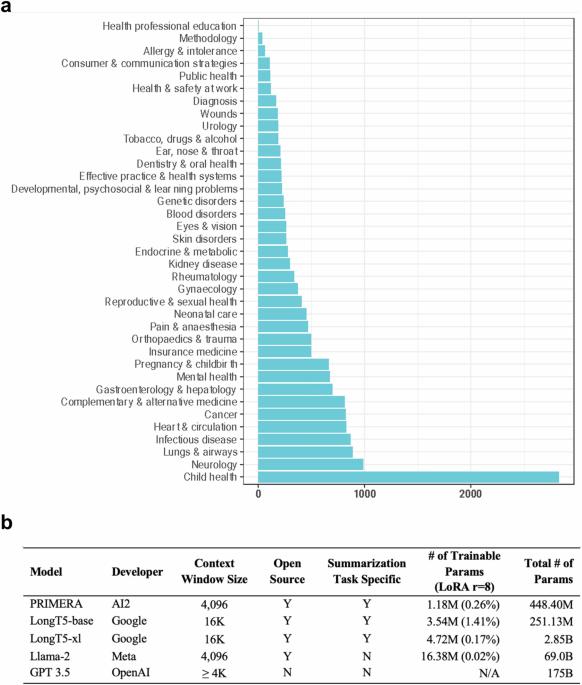
大型语言模型(LLM)在总结医学证据方面大有可为。最近的研究大多集中在专有 LLMs 的应用上。使用专有 LLM 会带来多种风险因素,包括缺乏透明度和对供应商的依赖性。虽然开源 LLMs 允许更好的透明度和定制化,但其性能却不及专有 LLMs。在本研究中,我们探讨了对开源 LLM 进行微调能在多大程度上进一步提高其性能。利用由 8161 对系统综述和摘要组成的基准数据集 MedReview,我们对 PRIMERA、LongT5 和 Llama-2 这三种广泛使用的开源 LLM 进行了微调。总体而言,开源模型的性能在微调后都有所提高。微调后的 LongT5 的性能接近零镜头设置下的 GPT-3.5。此外,较小的微调模型有时甚至比较大的零点模型表现出更优越的性能。上述改进趋势在人类评估和更大规模的 GPT4 模拟评估中都有所体现。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Closing the gap between open source and commercial large language models for medical evidence summarization
Large language models (LLMs) hold great promise in summarizing medical evidence. Most recent studies focus on the application of proprietary LLMs. Using proprietary LLMs introduces multiple risk factors, including a lack of transparency and vendor dependency. While open-source LLMs allow better transparency and customization, their performance falls short compared to the proprietary ones. In this study, we investigated to what extent fine-tuning open-source LLMs can further improve their performance. Utilizing a benchmark dataset, MedReview, consisting of 8161 pairs of systematic reviews and summaries, we fine-tuned three broadly-used, open-sourced LLMs, namely PRIMERA, LongT5, and Llama-2. Overall, the performance of open-source models was all improved after fine-tuning. The performance of fine-tuned LongT5 is close to GPT-3.5 with zero-shot settings. Furthermore, smaller fine-tuned models sometimes even demonstrated superior performance compared to larger zero-shot models. The above trends of improvement were manifested in both a human evaluation and a larger-scale GPT4-simulated evaluation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

NPJ Digital Medicine
Multiple-
CiteScore
25.10
自引率
3.30%
发文量
170
审稿时长
15 weeks
期刊介绍:
npj Digital Medicine is an online open-access journal that focuses on publishing peer-reviewed research in the field of digital medicine. The journal covers various aspects of digital medicine, including the application and implementation of digital and mobile technologies in clinical settings, virtual healthcare, and the use of artificial intelligence and informatics.
The primary goal of the journal is to support innovation and the advancement of healthcare through the integration of new digital and mobile technologies. When determining if a manuscript is suitable for publication, the journal considers four important criteria: novelty, clinical relevance, scientific rigor, and digital innovation.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


