利用基于生成式人工智能的虚拟多重肿瘤特征分析加速组织病理学工作流程
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
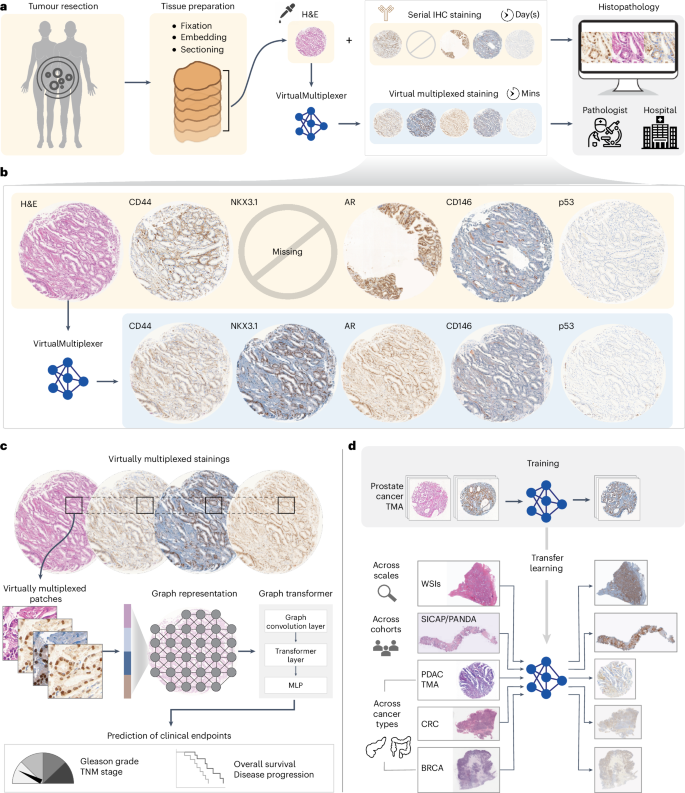
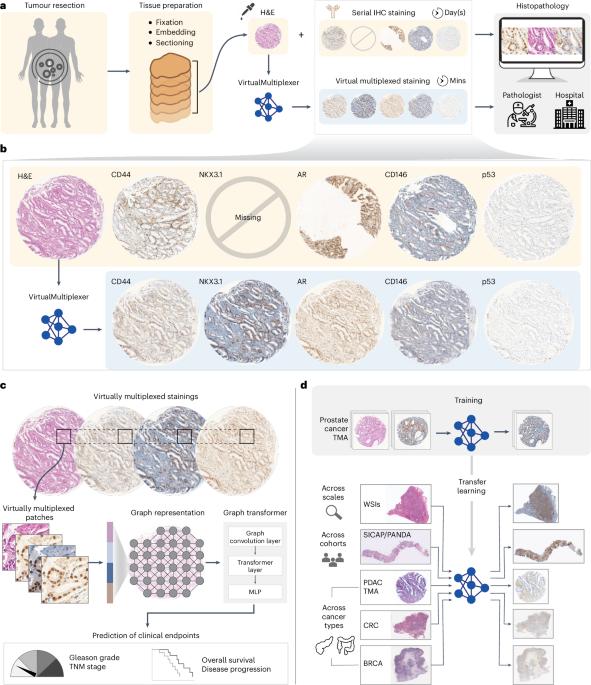
了解肿瘤的空间异质性及其与疾病发生和发展的关系是癌症生物学的基石。目前,组织病理学工作流程严重依赖苏木精、伊红和连续免疫组化染色,这是一个繁琐的组织排查过程,会导致组织图像不对齐。我们提出的 VirtualMultiplexer 是一种生成式人工智能工具包,它能从输入的苏木精和伊红图像中有效合成多个抗体标记物(即 AR、NKX3.1、CD44、CD146、p53 和 ERG)的多重免疫组化图像。VirtualMultiplexer 可捕捉跨组织尺度的生物相关染色模式,而无需连续的组织切片、图像注册或大量的专家注释。全面的定性和定量评估表明,VirtualMultiplexer 能够快速、稳健、精确地生成染色质量高且与真实染色无异的虚拟多重成像数据集。虚拟多路复用器可成功跨组织尺度和患者群组转移,无需对模型进行微调。最重要的是,虚拟多路复用图像能够训练图转换器,该转换器可同时从多个蛋白质的联合空间分布中学习,以预测临床相关终点。我们观察到,这种多重学习方案能够大大提高临床预测能力,这一点在多个下游任务、独立患者群和癌症类型中都得到了证实。我们的研究结果展示了人工智能辅助多路复用肿瘤成像的临床意义,加快了组织病理学工作流程和癌症生物学的发展。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Accelerating histopathology workflows with generative AI-based virtually multiplexed tumour profiling
Understanding the spatial heterogeneity of tumours and its links to disease initiation and progression is a cornerstone of cancer biology. Presently, histopathology workflows heavily rely on hematoxylin and eosin and serial immunohistochemistry staining, a cumbersome, tissue-exhaustive process that results in non-aligned tissue images. We propose the VirtualMultiplexer, a generative artificial intelligence toolkit that effectively synthesizes multiplexed immunohistochemistry images for several antibody markers (namely AR, NKX3.1, CD44, CD146, p53 and ERG) from only an input hematoxylin and eosin image. The VirtualMultiplexer captures biologically relevant staining patterns across tissue scales without requiring consecutive tissue sections, image registration or extensive expert annotations. Thorough qualitative and quantitative assessment indicates that the VirtualMultiplexer achieves rapid, robust and precise generation of virtually multiplexed imaging datasets of high staining quality that are indistinguishable from the real ones. The VirtualMultiplexer is successfully transferred across tissue scales and patient cohorts with no need for model fine-tuning. Crucially, the virtually multiplexed images enabled training a graph transformer that simultaneously learns from the joint spatial distribution of several proteins to predict clinically relevant endpoints. We observe that this multiplexed learning scheme was able to greatly improve clinical prediction, as corroborated across several downstream tasks, independent patient cohorts and cancer types. Our results showcase the clinical relevance of artificial intelligence-assisted multiplexed tumour imaging, accelerating histopathology workflows and cancer biology. VirtualMultiplexer is a generative AI tool that produces realistic multiplexed immunohistochemistry images from tissue biopsies. The generated images could be used to improve clinical predictions, enhancing histopathology workflows and accelerating cancer research.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


