利用全血表观基因组 DNA 甲基化数据开发结直肠癌生存预测模型。
IF 6.8
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
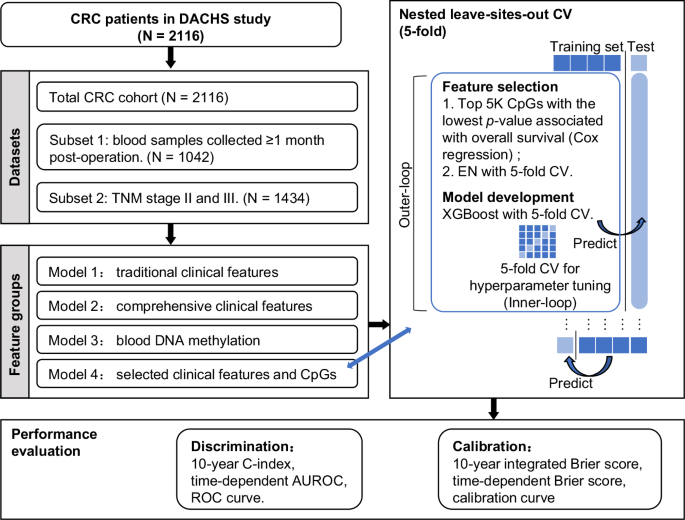
虽然全基因组关联研究在确定 CRC 生存预测因子方面很有价值,但将血液 DNA 甲基化(blood-DNAm)添加到临床特征(包括 TNM 系统)中的益处仍不清楚。在一项对2116名基线血液DNAm的CRC患者进行的多地点人群队列研究中,我们使用eXtreme Gradient Boosting技术分析了传统临床特征、综合临床特征、血液DNAm及其组合这四组患者的生存预测,并进行了5倍嵌套留点交叉验证。使用随时间变化的 ROC 曲线和校准来评估模型性能。在中位 10.3 年的随访期间,有 1166 名患者死亡。虽然基于血液 DNAm 的预测特征表现一般,但基于临床特征的预测特征优于基于血液 DNAm 的特征。与临床特征相比,加入血液 DNAm 并不能提高生存预测能力。M1分期、采血年龄和N2分期的贡献最大。尽管血液DNA甲基化具有一定的预后价值,但与临床特征相比,血液DNA甲基化并不能提高CRC患者的生存预测能力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Developing survival prediction models in colorectal cancer using epigenome-wide DNA methylation data from whole blood
While genome-wide association studies are valuable in identifying CRC survival predictors, the benefit of adding blood DNA methylation (blood-DNAm) to clinical features, including the TNM system, remains unclear. In a multi-site population-based patient cohort study of 2116 CRC patients with baseline blood-DNAm, we analyzed survival predictions using eXtreme Gradient Boosting with a 5-fold nested leave-sites-out cross-validation across four groups: traditional and comprehensive clinical features, blood-DNAm, and their combination. Model performance was assessed using time-dependent ROC curves and calibrations. During a median follow-up of 10.3 years, 1166 patients died. Although blood-DNAm-based predictive signatures achieved moderate performances, predictive signatures based on clinical features outperformed blood-DNAm signatures. The inclusion of blood-DNAm did not improve survival prediction over clinical features. M1 stage, age at blood collection, and N2 stage were the top contributors. Despite some prognostic value, incorporating blood DNA methylation did not enhance survival prediction of CRC patients beyond clinical features.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

NPJ Precision Oncology
ONCOLOGY-
CiteScore
9.90
自引率
1.30%
发文量
87
审稿时长
18 weeks
期刊介绍:
Online-only and open access, npj Precision Oncology is an international, peer-reviewed journal dedicated to showcasing cutting-edge scientific research in all facets of precision oncology, spanning from fundamental science to translational applications and clinical medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


