基于质谱的 tRNA 新直接测序和多种 RNA 修饰的定量制图
IF 14.4
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
尽管下一代 RNA 测序技术(NGS)得到了广泛应用,但同时对多种 RNA 核苷酸修饰进行直接测序和定量制图仍是一项挑战。基于质谱(MS)的测序可以直接对所有 RNA 修饰进行测序,而不局限于特定的修饰,但它需要一个完美的 MS 梯子,而很少有 tRNA 可以提供这样的梯子。在这里,我们描述了一种 MS 梯度互补测序方法(MLC-Seq),它规避了完美梯度的要求,允许以单核苷酸精度对具有多种核苷酸修饰的全长异质细胞 tRNA 进行从头 MS 测序。基于 NGS 的方法会丢失 RNA 修饰信息,而 MLC-Seq 则不同,它保留了 RNA 序列多样性和修饰信息,揭示了新的详细 tRNA 几何修饰图谱及其在脱烷基化酶 AlkB 处理后的变化。它还可以与参考序列相结合,对总 tRNA 样品中的各种 tRNA 和修饰进行定量分析。MLC-Seq 能够系统、定量、定点地绘制 RNA 修饰图谱,揭示 tRNA 真正完整的信息含量。本文章由计算机程序翻译,如有差异,请以英文原文为准。
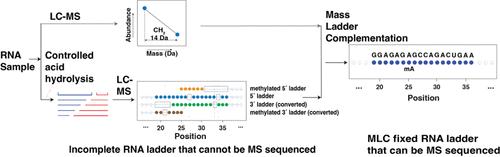
Mass Spectrometry-Based Direct Sequencing of tRNAs De Novo and Quantitative Mapping of Multiple RNA Modifications
Despite the extensive use of next-generation sequencing (NGS) of RNA, simultaneous direct sequencing and quantitative mapping of multiple RNA nucleotide modifications remains challenging. Mass spectrometry (MS)-based sequencing can directly sequence all RNA modifications without being limited to specific ones, but it requires a perfect MS ladder that few tRNAs can provide. Here, we describe an MS ladder complementation sequencing approach (MLC-Seq) that circumvents the perfect ladder requirement, allowing de novo MS sequencing of full-length heterogeneous cellular tRNAs with multiple nucleotide modifications at single-nucleotide precision. Unlike NGS-based methods, which lose RNA modification information, MLC-Seq preserves RNA sequence diversity and modification information, revealing new detailed stoichiometric tRNA modification profiles and their changes upon treatment with the dealkylating enzyme AlkB. It can also be combined with reference sequences to provide quantitative analysis of diverse tRNAs and modifications in total tRNA samples. MLC-Seq enables systematic, quantitative, and site-specific mapping of RNA modifications, revealing the truly complete informational content of tRNA.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
24.40
自引率
6.00%
发文量
2398
审稿时长
1.6 months
期刊介绍:
The flagship journal of the American Chemical Society, known as the Journal of the American Chemical Society (JACS), has been a prestigious publication since its establishment in 1879. It holds a preeminent position in the field of chemistry and related interdisciplinary sciences. JACS is committed to disseminating cutting-edge research papers, covering a wide range of topics, and encompasses approximately 19,000 pages of Articles, Communications, and Perspectives annually. With a weekly publication frequency, JACS plays a vital role in advancing the field of chemistry by providing essential research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


