{"title":"利用引物阻断非对称 PCR 进行单链 DNA 扩增的经济高效方法","authors":"Krisztina Percze, Ákos Harkai, Tamás Mészáros","doi":"10.1002/cpz1.1125","DOIUrl":null,"url":null,"abstract":"<p>In vitro amplification of single-stranded oligonucleotide libraries presents a significant challenge due to the potential for excessive byproduct formation. This phenomenon largely affects the quality of the ssDNAs created using the most commonly used methods, e.g., asymmetric PCR, biotin-streptavidin separation, or lambda exonuclease digestion of dsDNA. Here, we describe an improved protocol that combines primer-blocked asymmetric PCR (PBA-PCR) with emulsion PCR and a cost-effective downstream process that altogether alleviates byproduct formation without distorting the sequence space of the ssDNA library. In PBA-PCR, the reaction mixture is complemented with a 3′-phosphate-blocked limiting primer that decreases mispriming, thus reducing polymerization of DNA byproducts. The downstream process includes mixing of the PBA-PCR product with excess reverse complement of the 3′-phosphate-blocked limiting primer and removal of dsDNA strands via biotin-streptavidin separation, yielding purified ssDNAs. In conclusion, we have devised a universally applicable approach for simple and cost-effective production of ssDNA libraries and unique ssDNA sequences with on-demand labeling. Our protocol could be beneficial for a variety of uses, such as generating aptamer libraries for SELEX, creating unique molecular identifiers for a wide range of sequencing applications, providing donor DNA for CRISPR-Cas9 systems, developing scaffold nanostructures, and enabling DNA-based data storage. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Amplification of ssDNA libraries using PBA-PCR</p><p><b>Alternate Protocol 1</b>: Amplification of ssDNA libraries using emulsion PBA-PCR with a simplified extraction of PBA-PCR products</p><p><b>Basic Protocol 2</b>: Purification of PBA-PCR products to remove dsDNA and conversion of 3′-blocked primer to double-stranded complexes</p><p><b>Alternate Protocol 2</b>: Purification of PBA-PCR products to remove both dsDNA and blocking primers from the reaction mixture</p><p><b>Support Protocol</b>: Analysis of PBA-PCR products by gel electrophoresis</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 9","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-09-04","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1125","citationCount":"0","resultStr":"{\"title\":\"A Cost-Effective Approach for Single-Stranded DNA Amplification Using Primer-Blocked Asymmetric PCR\",\"authors\":\"Krisztina Percze, Ákos Harkai, Tamás Mészáros\",\"doi\":\"10.1002/cpz1.1125\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>In vitro amplification of single-stranded oligonucleotide libraries presents a significant challenge due to the potential for excessive byproduct formation. This phenomenon largely affects the quality of the ssDNAs created using the most commonly used methods, e.g., asymmetric PCR, biotin-streptavidin separation, or lambda exonuclease digestion of dsDNA. Here, we describe an improved protocol that combines primer-blocked asymmetric PCR (PBA-PCR) with emulsion PCR and a cost-effective downstream process that altogether alleviates byproduct formation without distorting the sequence space of the ssDNA library. In PBA-PCR, the reaction mixture is complemented with a 3′-phosphate-blocked limiting primer that decreases mispriming, thus reducing polymerization of DNA byproducts. The downstream process includes mixing of the PBA-PCR product with excess reverse complement of the 3′-phosphate-blocked limiting primer and removal of dsDNA strands via biotin-streptavidin separation, yielding purified ssDNAs. In conclusion, we have devised a universally applicable approach for simple and cost-effective production of ssDNA libraries and unique ssDNA sequences with on-demand labeling. Our protocol could be beneficial for a variety of uses, such as generating aptamer libraries for SELEX, creating unique molecular identifiers for a wide range of sequencing applications, providing donor DNA for CRISPR-Cas9 systems, developing scaffold nanostructures, and enabling DNA-based data storage. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Amplification of ssDNA libraries using PBA-PCR</p><p><b>Alternate Protocol 1</b>: Amplification of ssDNA libraries using emulsion PBA-PCR with a simplified extraction of PBA-PCR products</p><p><b>Basic Protocol 2</b>: Purification of PBA-PCR products to remove dsDNA and conversion of 3′-blocked primer to double-stranded complexes</p><p><b>Alternate Protocol 2</b>: Purification of PBA-PCR products to remove both dsDNA and blocking primers from the reaction mixture</p><p><b>Support Protocol</b>: Analysis of PBA-PCR products by gel electrophoresis</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":\"4 9\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-09-04\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1125\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1125\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1125","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
A Cost-Effective Approach for Single-Stranded DNA Amplification Using Primer-Blocked Asymmetric PCR
In vitro amplification of single-stranded oligonucleotide libraries presents a significant challenge due to the potential for excessive byproduct formation. This phenomenon largely affects the quality of the ssDNAs created using the most commonly used methods, e.g., asymmetric PCR, biotin-streptavidin separation, or lambda exonuclease digestion of dsDNA. Here, we describe an improved protocol that combines primer-blocked asymmetric PCR (PBA-PCR) with emulsion PCR and a cost-effective downstream process that altogether alleviates byproduct formation without distorting the sequence space of the ssDNA library. In PBA-PCR, the reaction mixture is complemented with a 3′-phosphate-blocked limiting primer that decreases mispriming, thus reducing polymerization of DNA byproducts. The downstream process includes mixing of the PBA-PCR product with excess reverse complement of the 3′-phosphate-blocked limiting primer and removal of dsDNA strands via biotin-streptavidin separation, yielding purified ssDNAs. In conclusion, we have devised a universally applicable approach for simple and cost-effective production of ssDNA libraries and unique ssDNA sequences with on-demand labeling. Our protocol could be beneficial for a variety of uses, such as generating aptamer libraries for SELEX, creating unique molecular identifiers for a wide range of sequencing applications, providing donor DNA for CRISPR-Cas9 systems, developing scaffold nanostructures, and enabling DNA-based data storage. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
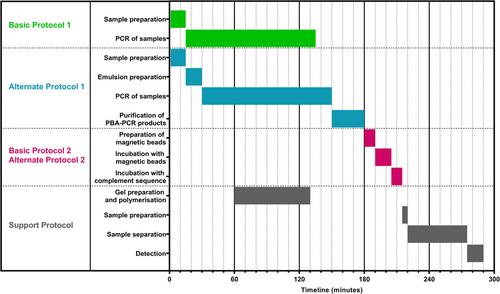
Basic Protocol 1: Amplification of ssDNA libraries using PBA-PCR
Alternate Protocol 1: Amplification of ssDNA libraries using emulsion PBA-PCR with a simplified extraction of PBA-PCR products
Basic Protocol 2: Purification of PBA-PCR products to remove dsDNA and conversion of 3′-blocked primer to double-stranded complexes
Alternate Protocol 2: Purification of PBA-PCR products to remove both dsDNA and blocking primers from the reaction mixture
Support Protocol: Analysis of PBA-PCR products by gel electrophoresis

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


