在血液恶性肿瘤中通过 RNA 测序初步检测到罕见融合转录本后,对其进行定制的数字 PCR 追踪。
IF 3.4
3区 医学
Q1 PATHOLOGY
引用次数: 0
摘要
血液恶性肿瘤的治疗已进入一个新时代,其中最小残留病(MRD)监测发挥着关键作用。PML::RARA、CBFB::MYH11 或 RUNX1::RUNX1T1等公认的分子靶标通常通过定量反转录 PCR 追踪。最近,通过转录组分析,人们发现了更广泛的融合转录本。这些新发现的融合转录本可能成为 MRD 定量的新型分子标记。在本研究中,我们比较了靶向 RNA-seq 方法(FusionPlex)和全转录组策略(Advanta RNA-seq XT)在 21 个样本训练集中的融合检测。在已知融合的检测中,我们发现两者的一致性达到了 100%,而且两种技术在基因表达定量方面显示出良好的相关性(Spearman r=0.77)。此外,我们还在 126 例血液恶性肿瘤患者中对通过靶向 RNA-seq 鉴定融合进行了前瞻性评估。60名患者(48%)至少检测到一个融合转录本。我们设计了针对 11 种罕见融合的定制数字 PCR 检测方法,并验证了该技术在 MRD 定量中的检测限低于 0.01%。RNA-seq 和定制数字 PCR 的结合可能成为缺乏常规分子靶点的患者进行 MRD 评估的新标准。本文章由计算机程序翻译,如有差异,请以英文原文为准。
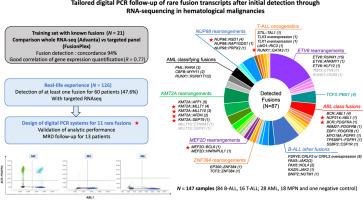
Tailored Digital PCR Follow-Up of Rare Fusion Transcripts after Initial Detection through RNA Sequencing in Hematological Malignancies
Minimal residual disease (MRD) monitoring plays a pivotal role in the management of hematologic malignancies. Well-established molecular targets, such as PML::RARA, CBFB::MYH11, or RUNX1::RUNX1T1, are conventionally tracked by quantitative RT-PCR. Recently, a broader landscape of fusion transcripts has been unveiled through transcriptomic analysis. These newly discovered fusion transcripts may emerge as novel molecular markers for MRD quantification. In this study, we compared a targeted RNA-sequencing (RNA-seq) approach (FusionPlex) with a whole-transcriptomic strategy (Advanta RNA-Seq XT) for fusion detection in a training set of 21 samples. We evidenced a concordance of 100% for the detection of known fusions, and showed a good correlation for gene expression quantification between the two techniques (Spearman r = 0.77). Additionally, we prospectively evaluated the identification of fusions by targeted RNA-seq in a real-life series of 126 patients with hematological malignancy. At least one fusion transcript was detected for 60 patients (48%). We designed tailored digital PCR assays for 11 rare fusions, and validated this technique for MRD quantification with a limit of detection of <0.01%. The combination of RNA-seq and tailored digital PCR may become a new standard for MRD evaluation in patients lacking conventional molecular targets.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
8.10
自引率
2.40%
发文量
143
审稿时长
43 days
期刊介绍:
The Journal of Molecular Diagnostics, the official publication of the Association for Molecular Pathology (AMP), co-owned by the American Society for Investigative Pathology (ASIP), seeks to publish high quality original papers on scientific advances in the translation and validation of molecular discoveries in medicine into the clinical diagnostic setting, and the description and application of technological advances in the field of molecular diagnostic medicine. The editors welcome for review articles that contain: novel discoveries or clinicopathologic correlations including studies in oncology, infectious diseases, inherited diseases, predisposition to disease, clinical informatics, or the description of polymorphisms linked to disease states or normal variations; the application of diagnostic methodologies in clinical trials; or the development of new or improved molecular methods which may be applied to diagnosis or monitoring of disease or disease predisposition.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


