IMPRESS:利用限制性酶和 smMIP 测序改进甲基化分析,并结合新的生物标记物面板,创建多种癌症检测分析。
IF 6.4
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
背景:尽管癌症诊断在全球范围内取得了进展,但仍需要更灵敏的诊断生物标志物。在过去的几十年中,人们对甲基组进行了广泛的研究,但仍缺乏一种用于多重分析的低成本、不含亚硫酸氢盐的检测方法:方法:我们开发了一种名为 IMPRESS 的甲基化检测技术,它结合了甲基化敏感限制酶和单分子分子反转探针。我们利用这种技术开发了一种多癌症检测试剂盒,用于检测全球八种致死率最高的癌症类型。我们选择了 1791 个可根据 DNA 甲基化区分肿瘤和正常组织的 CpG 位点。我们用 IMPRESS 分析了 35 份血液样本、111 份肿瘤样本和 114 份正常样本中的这些位点。最后,建立了一个分类器模型:我们介绍了 IMPRESS 的成功开发,并用 ddPCR 对其进行了验证。最终建立的分类器模型可通过 358 个 CpG 靶点区分肿瘤和正常样本,灵敏度为 0.95,特异度为 0.91。此外,我们提供的数据还凸显了 IMPRESS 在液体活检方面的潜力:我们成功地创建了一种创新的 DNA 甲基化检测技术。结论:我们成功地创造了一种创新的 DNA 甲基化检测技术,通过将这种方法与新的多癌症生物标记物面板相结合,我们开发出了一种灵敏而特异的多癌症检测方法,有望用于液体活检。本文章由计算机程序翻译,如有差异,请以英文原文为准。
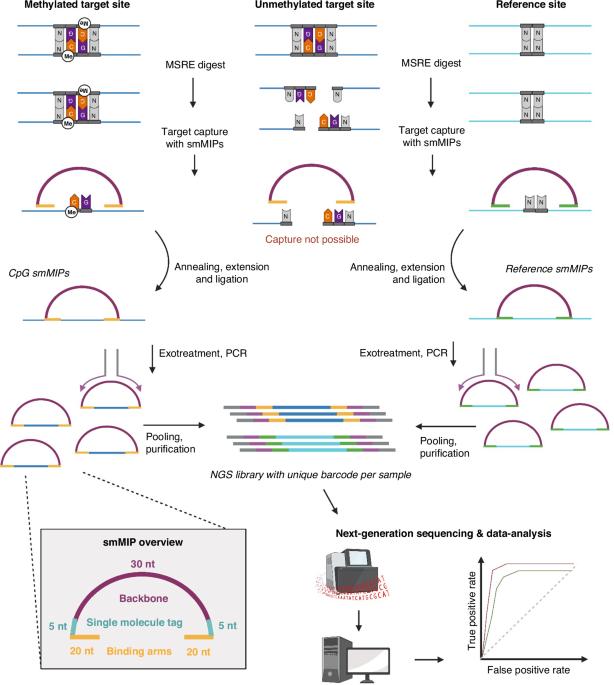
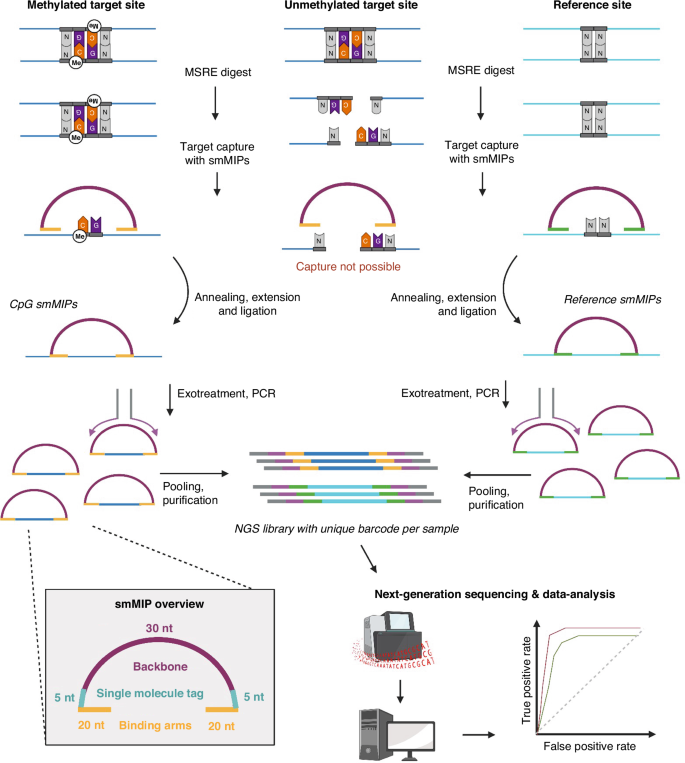
IMPRESS: Improved methylation profiling using restriction enzymes and smMIP sequencing, combined with a new biomarker panel, creating a multi-cancer detection assay
Despite the worldwide progress in cancer diagnostics, more sensitive diagnostic biomarkers are needed. The methylome has been extensively investigated in the last decades, but a low-cost, bisulfite-free detection method for multiplex analysis is still lacking. We developed a methylation detection technique called IMPRESS, which combines methylation-sensitive restriction enzymes and single-molecule Molecular Inversion Probes. We used this technique for the development of a multi-cancer detection assay for eight of the most lethal cancer types worldwide. We selected 1791 CpG sites that can distinguish tumor from normal tissue based on DNA methylation. These sites were analysed with IMPRESS in 35 blood, 111 tumor and 114 normal samples. Finally, a classifier model was built. We present the successful development of IMPRESS and validated it with ddPCR. The final classifier model discriminating tumor from normal samples was built with 358 CpG target sites and reached a sensitivity of 0.95 and a specificity of 0.91. Moreover, we provide data that highlight IMPRESS’s potential for liquid biopsies. We successfully created an innovative DNA methylation detection technique. By combining this method with a new multi-cancer biomarker panel, we developed a sensitive and specific multi-cancer assay, with potential use in liquid biopsies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

British Journal of Cancer
医学-肿瘤学
CiteScore
15.10
自引率
1.10%
发文量
383
审稿时长
6 months
期刊介绍:
The British Journal of Cancer is one of the most-cited general cancer journals, publishing significant advances in translational and clinical cancer research.It also publishes high-quality reviews and thought-provoking comment on all aspects of cancer prevention,diagnosis and treatment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


