综合耐药性和白血病干细胞基因表达评分可预测来自 10 项试验的 3500 多名急性髓细胞性白血病患者的预后。
IF 6.8
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
在本研究中,我们利用机器学习工具,评估了儿科急性髓细胞性白血病患者发现队列(N = 163;NCT00136084)中与标准急性髓细胞性白血病化疗(阿糖胞苷/大柔比星/依托泊苷)药理学相关的基因表达,并定义了可预测预后的 5 基因耐药评分(ADE-RS5)(高 MRD1 阳性 p = 0.013;较低 EFS p = 0.013;较低 EFS p = 0.013)。本文章由计算机程序翻译,如有差异,请以英文原文为准。
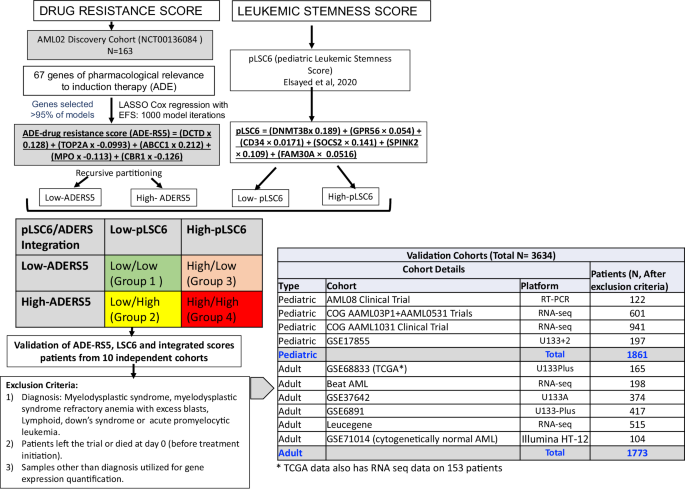
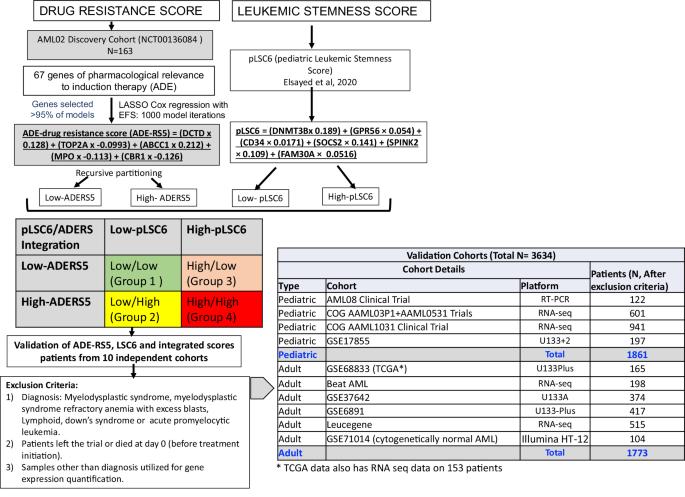
Integrated drug resistance and leukemic stemness gene-expression scores predict outcomes in large cohort of over 3500 AML patients from 10 trials
In this study, we leveraged machine-learning tools by evaluating expression of genes of pharmacological relevance to standard-AML chemotherapy (ara-C/daunorubicin/etoposide) in a discovery-cohort of pediatric AML patients (N = 163; NCT00136084 ) and defined a 5-gene-drug resistance score (ADE-RS5) that was predictive of outcome (high MRD1 positivity p = 0.013; lower EFS p < 0.0001 and OS p < 0.0001). ADE-RS5 was integrated with a previously defined leukemic-stemness signature (pLSC6) to classify patients into four groups. ADE-RS5, pLSC6 and integrated-score was evaluated for association with outcome in one of the largest assembly of ~3600 AML patients from 10 independent cohorts (1861 pediatric and 1773 adult AML). Patients with high ADE-RS5 had poor outcome in validation cohorts and the previously reported pLSC6 maintained strong significant association in all validation cohorts. For pLSC6/ADE-RS5-integrated-score analysis, using Group-1 (low-scores for ADE-RS5 and pLSC6) as reference, Group-4 (high-scores for ADE-RS5 and pLSC6) showed worst outcome (EFS: p < 0.0001 and OS: p < 0.0001). Groups-2/3 (one high and one low-score) showed intermediate outcome (p < 0.001). Integrated score groups remained an independent predictor of outcome in multivariable-analysis after adjusting for established prognostic factors (EFS: Group 2 vs. 1, HR = 4.68, p < 0.001, Group 3 vs. 1, HR = 3.22, p = 0.01, and Group 4 vs. 1, HR = 7.26, p < 0.001). These results highlight the significant prognostic value of transcriptomics-based scores capturing disease aggressiveness through pLSC6 and drug resistance via ADE-RS5. The pLSC6 stemness score is a significant predictor of outcome and associates with high-risk group features, the ADE-RS5 drug resistance score adds further value, reflecting the clinical utility of simultaneous testing of both for optimizing treatment strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

NPJ Precision Oncology
ONCOLOGY-
CiteScore
9.90
自引率
1.30%
发文量
87
审稿时长
18 weeks
期刊介绍:
Online-only and open access, npj Precision Oncology is an international, peer-reviewed journal dedicated to showcasing cutting-edge scientific research in all facets of precision oncology, spanning from fundamental science to translational applications and clinical medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


