揭示乙型肝炎病毒 DNA 整合在 B 细胞淋巴瘤形成中的作用。
IF 6.4
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
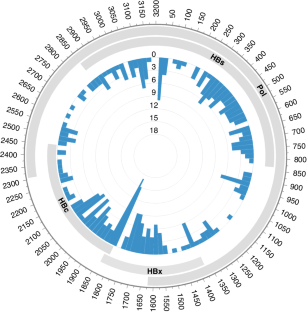
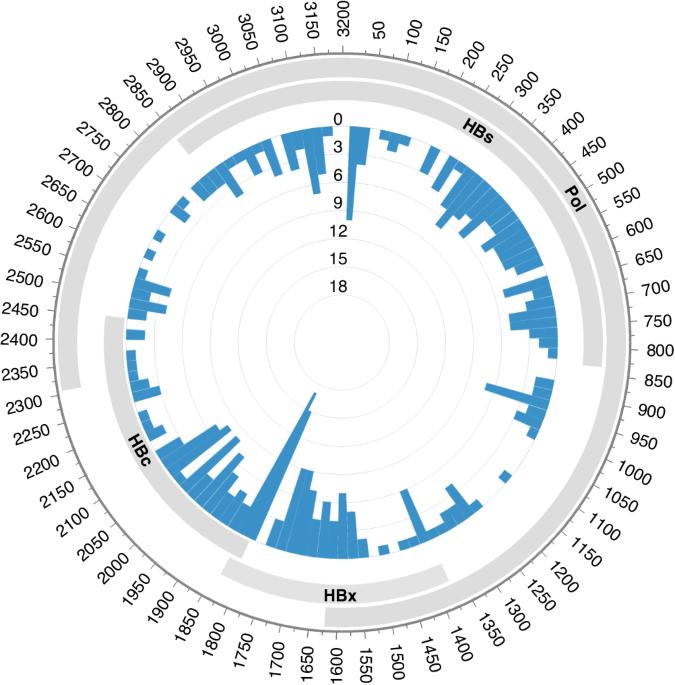
背景:研究表明,乙型肝炎病毒(HBV)相关的B细胞非霍奇金淋巴瘤(NHL)是一个独特的亚组,具有不同的临床特征。HBV DNA整合到B细胞基因组中是否是淋巴瘤发病的一个因果机制,这个问题仍然悬而未决:方法:我们使用基于杂交捕获的新一代测序技术和 RNA 测序技术,分析了 45 例 HBV 相关 B 细胞 NHL 肿瘤组织中的 HBV 整合模式:结果:在13个样本(28.9%)中共发现了354个HBV整合位点,表明B细胞NHL中的整合频率相对较低。高血浆 HBV DNA 负荷与 HBV 整合的存在无关。插入位点随机分布在淋巴瘤的所有基因组中,在染色体水平和基因水平上都没有任何偏好热点。耐人寻味的是,在B细胞NHL中,大多数HBV整合是非克隆性的,这意味着它们不会带来生存优势。对诊断-复发配对样本的分析表明,在疾病进展过程中,HBV整合的状态并不稳定。此外,转录组分析表明,HBV整合的生物学影响有限:我们的研究提供了B细胞NHL中无偏见的HBV整合图谱,揭示了HBV DNA整合在B细胞淋巴瘤发生中的微不足道的作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Unraveling the role of hepatitis B virus DNA integration in B-cell lymphomagenesis
Studies have shown that hepatitis B virus (HBV)-associated B-cell non-Hodgkin lymphoma (NHL) constitutes a unique subgroup with distinct clinical features. It still leaves open the question of whether the integration of HBV DNA into the B-cell genome is a causal mechanism in the development of lymphoma. Using the hybridisation capture-based next generation sequencing and RNA sequencing, we characterised the HBV integration pattern in 45 HBV-associated B-cell NHL tumour tissues. A total of 354 HBV integration sites were identified in 13 (28.9%) samples, indicating the relatively low integration frequency in B-cell NHLs. High plasma HBV DNA loads were not associated with the existence of HBV integration. The insertion sites distributed randomly across all the lymphoma genome without any preferential hotspot neither at the chromosomal level nor at the genetic level. Intriguingly, most HBV integrations were nonclonal in B-cell NHLs, implying that they did not confer a survival advantage. Analysis of the paired diagnosis-relapse samples showed the unstable status of HBV integrations during disease progression. Furthermore, transcriptomic analysis revealed the limited biological impact of HBV integration. Our study provides an unbiased HBV integration map in B-cell NHLs, revealing the insignificant role of HBV DNA integration in B-cell lymphomagenesis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

British Journal of Cancer
医学-肿瘤学
CiteScore
15.10
自引率
1.10%
发文量
383
审稿时长
6 months
期刊介绍:
The British Journal of Cancer is one of the most-cited general cancer journals, publishing significant advances in translational and clinical cancer research.It also publishes high-quality reviews and thought-provoking comment on all aspects of cancer prevention,diagnosis and treatment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


