利用神经场网络对低温电子显微镜结构进行高分辨率实空间重建
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
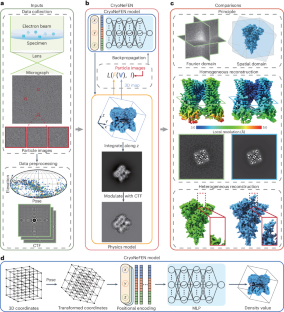
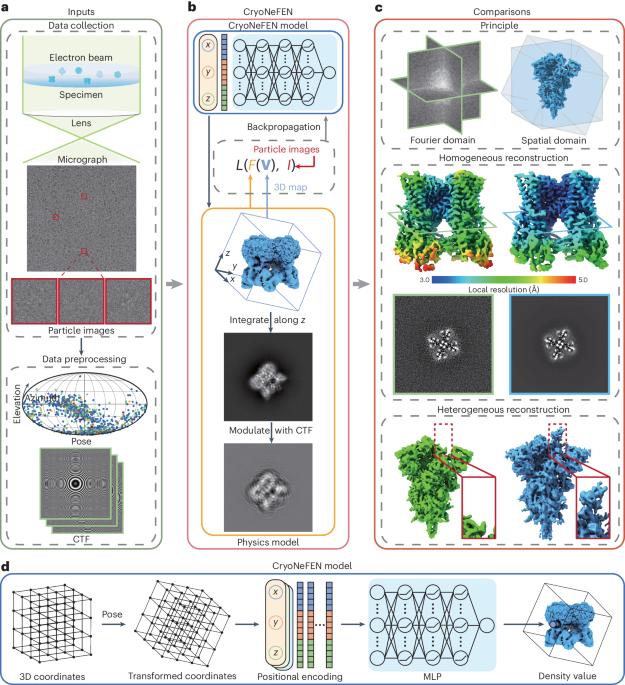
阐明三维(3D)结构对于揭示蛋白质功能和阐明结构生物学机制至关重要。低温电子显微镜(cryo-EM)单颗粒分析为确定大分子结构提供了直接测量方法。然而,从噪声极高且随机定向的二维投影图像中重建高分辨率三维结构是一大挑战。现有方法大多涉及傅立叶域中多个二维切片的优化,但忽略了这些切片之间的各向异性,从而限制了高频结构的重建。在本文中,我们提出了一种利用三维空间域优化的冷冻电镜神经场重建网络,该网络通过将空间坐标映射到相应的密度值来学习冷冻电镜结构的定向各向同性表示。我们在四个数据集上对低温电磁神经场重建网络进行了定性和定量评估。在同质重建中,冷冻电镜神经场重建网络提高了方向各向同性和三维密度分辨率,超越了现有算法的极限,并在异质重建中解决了 SARS-CoV-2 缺失的元素。本文章由计算机程序翻译,如有差异,请以英文原文为准。
High-resolution real-space reconstruction of cryo-EM structures using a neural field network
The elucidation of three-dimensional (3D) structures is crucial for unravelling the protein function and illuminating mechanisms in structural biology. Cryogenic electron microscopy (cryo-EM) single-particle analysis provides direct measurements to determine the structures of macromolecules. However, the main challenge is reconstructing high-resolution 3D structures from extremely noisy and randomly oriented two-dimensional projection images. Most existing methods involve the optimization of multiple two-dimensional slices in the Fourier domain but ignore the anisotropy among these slices, thereby limiting the reconstruction of high-frequency structures. In this paper, we propose a cryo-EM neural field reconstruction network using 3D spatial-domain optimization that learns a directional isotropic representation of the cryo-EM structure by mapping the spatial coordinates to the corresponding density values. We qualitatively and quantitatively evaluate the cryo-EM neural field reconstruction network on four datasets. The cryo-EM neural field reconstruction network improves the directional isotropy and 3D density resolution beyond the limits of existing algorithms in homogeneous reconstruction and resolves the missing elements of SARS-CoV-2 in heterogeneous reconstruction. Elucidating three-dimensional structures is crucial for unravelling the macromolecule function in structural biology. This study presents a cryogenic electron microscopy neural field reconstruction network using real-space optimization, enhancing the resolution in cryogenic electron microscopy reconstruction.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


