Bart van der Sanden, Kornelia Neveling, Andy Wing Chun Pang, Syukri Shukor, Michael D. Gallagher, Stephanie L. Burke, Erik-Jan Kamsteeg, Alex Hastie, Alexander Hoischen
下载PDF
{"title":"应用于重复扩展疾病的光学基因组图谱。","authors":"Bart van der Sanden, Kornelia Neveling, Andy Wing Chun Pang, Syukri Shukor, Michael D. Gallagher, Stephanie L. Burke, Erik-Jan Kamsteeg, Alex Hastie, Alexander Hoischen","doi":"10.1002/cpz1.1094","DOIUrl":null,"url":null,"abstract":"<p>Short tandem repeat (STR) expansions are associated with more than 60 genetic disorders. The size and stability of these expansions correlate with the severity and age of onset of the disease. Therefore, being able to accurately detect the absolute length of STRs is important. Current diagnostic assays include laborious lab experiments, including repeat-primed PCR and Southern blotting, that still cannot precisely determine the exact length of very long repeat expansions. Optical genome mapping (OGM) is a cost-effective and easy-to-use alternative to traditional cytogenetic techniques and allows the comprehensive detection of chromosomal aberrations and structural variants >500 bp in length, including insertions, deletions, duplications, inversions, translocations, and copy number variants. Here, we provide methodological guidance for preparing samples and performing OGM as well as running the analysis pipelines and using the specific repeat expansion workflows to determine the exact repeat length of repeat expansions expanded beyond 500 bp. Together these protocols provide all details needed to analyze the length and stability of any repeat expansion with an expected repeat size difference from the expected wild-type allele of >500 bp. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Genomic ultra-high-molecular-weight DNA isolation, labeling, and staining</p><p><b>Basic Protocol 2</b>: Data generation and genome mapping using the Bionano Saphyr® System</p><p><b>Basic Protocol 3</b>: Manual <i>De Novo</i> Assembly workflow</p><p><b>Basic Protocol 4</b>: Local guided assembly workflow</p><p><b>Basic Protocol 5</b>: EnFocus Fragile X workflow</p><p><b>Basic Protocol 6</b>: Molecule distance script workflow</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 7","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-07-05","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1094","citationCount":"0","resultStr":"{\"title\":\"Optical Genome Mapping for Applications in Repeat Expansion Disorders\",\"authors\":\"Bart van der Sanden, Kornelia Neveling, Andy Wing Chun Pang, Syukri Shukor, Michael D. Gallagher, Stephanie L. Burke, Erik-Jan Kamsteeg, Alex Hastie, Alexander Hoischen\",\"doi\":\"10.1002/cpz1.1094\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Short tandem repeat (STR) expansions are associated with more than 60 genetic disorders. The size and stability of these expansions correlate with the severity and age of onset of the disease. Therefore, being able to accurately detect the absolute length of STRs is important. Current diagnostic assays include laborious lab experiments, including repeat-primed PCR and Southern blotting, that still cannot precisely determine the exact length of very long repeat expansions. Optical genome mapping (OGM) is a cost-effective and easy-to-use alternative to traditional cytogenetic techniques and allows the comprehensive detection of chromosomal aberrations and structural variants >500 bp in length, including insertions, deletions, duplications, inversions, translocations, and copy number variants. Here, we provide methodological guidance for preparing samples and performing OGM as well as running the analysis pipelines and using the specific repeat expansion workflows to determine the exact repeat length of repeat expansions expanded beyond 500 bp. Together these protocols provide all details needed to analyze the length and stability of any repeat expansion with an expected repeat size difference from the expected wild-type allele of >500 bp. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Genomic ultra-high-molecular-weight DNA isolation, labeling, and staining</p><p><b>Basic Protocol 2</b>: Data generation and genome mapping using the Bionano Saphyr® System</p><p><b>Basic Protocol 3</b>: Manual <i>De Novo</i> Assembly workflow</p><p><b>Basic Protocol 4</b>: Local guided assembly workflow</p><p><b>Basic Protocol 5</b>: EnFocus Fragile X workflow</p><p><b>Basic Protocol 6</b>: Molecule distance script workflow</p>\",\"PeriodicalId\":93970,\"journal\":{\"name\":\"Current protocols\",\"volume\":\"4 7\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-07-05\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.1094\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1094\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.1094","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
引用
批量引用
Optical Genome Mapping for Applications in Repeat Expansion Disorders
Short tandem repeat (STR) expansions are associated with more than 60 genetic disorders. The size and stability of these expansions correlate with the severity and age of onset of the disease. Therefore, being able to accurately detect the absolute length of STRs is important. Current diagnostic assays include laborious lab experiments, including repeat-primed PCR and Southern blotting, that still cannot precisely determine the exact length of very long repeat expansions. Optical genome mapping (OGM) is a cost-effective and easy-to-use alternative to traditional cytogenetic techniques and allows the comprehensive detection of chromosomal aberrations and structural variants >500 bp in length, including insertions, deletions, duplications, inversions, translocations, and copy number variants. Here, we provide methodological guidance for preparing samples and performing OGM as well as running the analysis pipelines and using the specific repeat expansion workflows to determine the exact repeat length of repeat expansions expanded beyond 500 bp. Together these protocols provide all details needed to analyze the length and stability of any repeat expansion with an expected repeat size difference from the expected wild-type allele of >500 bp. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
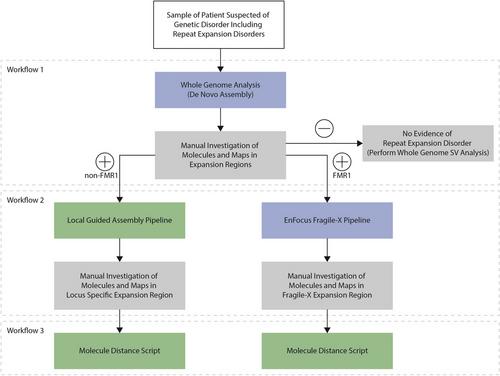
Basic Protocol 1 : Genomic ultra-high-molecular-weight DNA isolation, labeling, and staining
Basic Protocol 2 : Data generation and genome mapping using the Bionano Saphyr® System
Basic Protocol 3 : Manual De Novo Assembly workflow
Basic Protocol 4 : Local guided assembly workflow
Basic Protocol 5 : EnFocus Fragile X workflow
Basic Protocol 6 : Molecule distance script workflow

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


