通过整合物理先验知识和数据增强建模进行通用蛋白质配体相互作用评分
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
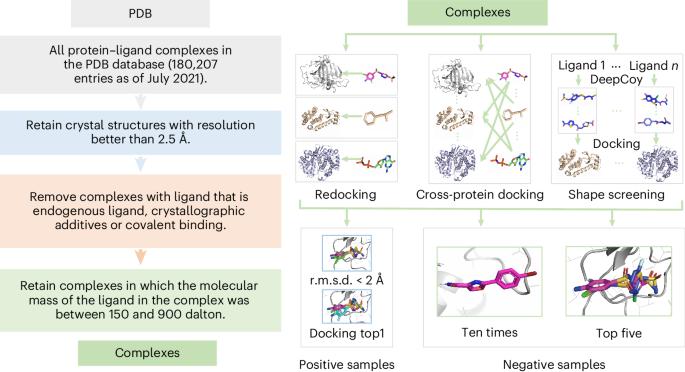
开发稳健的蛋白质配体相互作用评估方法是一个长期存在的问题。数据驱动的方法可能会记忆配体和蛋白质的训练数据,而不是学习蛋白质配体之间的相互作用。在这里,我们展示了一种名为 EquiScore 的评分方法,它利用异构图神经网络整合物理先验知识,在等变几何空间中描述蛋白质-配体的相互作用。EquiScore 基于一个新的数据集进行训练,该数据集采用了多种数据增强策略和严格的冗余去除方案。在两个大型外部测试集上,与其他 21 种方法相比,EquiScore 的性能始终名列前茅。当 EquiScore 与不同的对接方法一起使用时,它能有效地提高这些对接方法的筛选能力。EquiScore 还在一系列结构类似物的活性排名任务中表现出良好的性能,这表明它具有指导先导化合物优化的潜力。最后,我们研究了 EquiScore 不同层次的可解释性,这可能会为基于结构的药物设计提供更多启示。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Generic protein–ligand interaction scoring by integrating physical prior knowledge and data augmentation modelling
Developing robust methods for evaluating protein–ligand interactions has been a long-standing problem. Data-driven methods may memorize ligand and protein training data rather than learning protein–ligand interactions. Here we show a scoring approach called EquiScore, which utilizes a heterogeneous graph neural network to integrate physical prior knowledge and characterize protein–ligand interactions in equivariant geometric space. EquiScore is trained based on a new dataset constructed with multiple data augmentation strategies and a stringent redundancy-removal scheme. On two large external test sets, EquiScore consistently achieved top-ranking performance compared to 21 other methods. When EquiScore is used alongside different docking methods, it can effectively enhance the screening ability of these docking methods. EquiScore also showed good performance on the activity-ranking task of a series of structural analogues, indicating its potential to guide lead compound optimization. Finally, we investigated different levels of interpretability of EquiScore, which may provide more insights into structure-based drug design. Machine learning can improve scoring methods to evaluate protein–ligand interactions, but achieving good generalization is an outstanding challenge. Cao et al. introduce EquiScore, which is based on a graph neural network that integrates physical knowledge and is shown to have robust capabilities when applied to unseen protein targets.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


