揭示急性髓性白血病的线粒体和核糖体基因失调及肿瘤微环境动态。
IF 4.8
3区 医学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
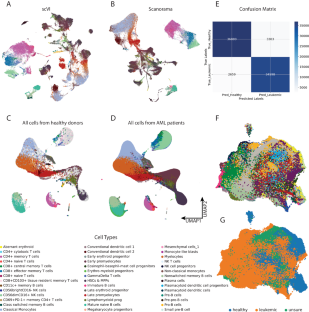
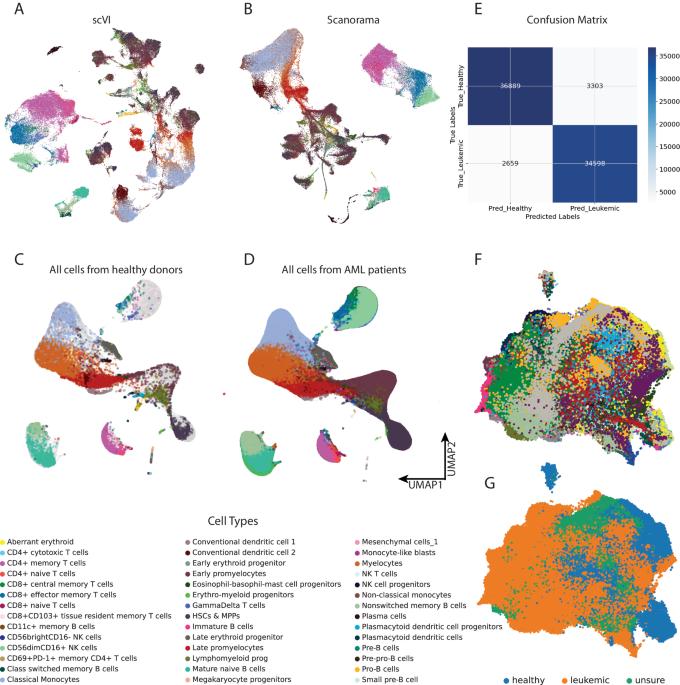
急性髓性白血病(AML)是一种预后不良的恶性克隆性造血疾病。了解白血病细胞与肿瘤微环境(TME)之间的相互作用有助于预测白血病的预后并指导治疗。使用基于Python的管道(包括基于机器学习的scVI工具)重新分析来自CSC和G20队列的scRNA-seq数据,再现了急性髓细胞性白血病患者样本中独特的分层结构。通过加权相关网络分析(WGCNA)构建了加权基因共表达网络,并确定了主要集中于造血干细胞(HSCs)、多能祖细胞(MPPs)和自然杀伤细胞(NK)的基因模块。分析显示,与有氧呼吸和核糖体/细胞质翻译相关的基因模块出现了明显的失调。CellChat 软件包阐明了细胞与细胞之间的交流,揭示了激活性和抑制性免疫信号通路的失衡。截取白血病造血干细胞和骨髓造血干细胞以及高NKG2A的NK细胞中上调的基因用于构建预后模型。基于 10 个基因建立了正常 Cox 模型和人工神经网络模型。该研究揭示了急性髓细胞性白血病患者线粒体和核糖体基因的失调,并提示急性髓细胞性白血病TME中同时存在刺激和抑制因子。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Unveiling mitochondrial and ribosomal gene deregulation and tumor microenvironment dynamics in acute myeloid leukemia
Acute myeloid leukemia (AML) is a malignant clonal hematopoietic disease with a poor prognosis. Understanding the interaction between leukemic cells and the tumor microenvironment (TME) can help predict the prognosis of leukemia and guide its treatment. Re-analyzing the scRNA-seq data from the CSC and G20 cohorts, using a Python-based pipeline including machine-learning-based scVI-tools, recapitulated the distinct hierarchical structure within the samples of AML patients. Weighted correlation network analysis (WGCNA) was conducted to construct a weighted gene co-expression network and to identify gene modules primarily focusing on hematopoietic stem cells (HSCs), multipotent progenitors (MPPs), and natural killer (NK) cells. The analysis revealed significant deregulation in gene modules associated with aerobic respiration and ribosomal/cytoplasmic translation. Cell–cell communications were elucidated by the CellChat package, revealing an imbalance of activating and inhibitory immune signaling pathways. Interception of genes upregulated in leukemic HSCs & MPPs as well as in NKG2A-high NK cells was used to construct prognostic models. Normal Cox and artificial neural network models based on 10 genes were developed. The study reveals the deregulation of mitochondrial and ribosomal genes in AML patients and suggests the co-occurrence of stimulatory and inhibitory factors in the AML TME.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cancer gene therapy
医学-生物工程与应用微生物
CiteScore
10.20
自引率
0.00%
发文量
150
审稿时长
4-8 weeks
期刊介绍:
Cancer Gene Therapy is the essential gene and cellular therapy resource for cancer researchers and clinicians, keeping readers up to date with the latest developments in gene and cellular therapies for cancer. The journal publishes original laboratory and clinical research papers, case reports and review articles. Publication topics include RNAi approaches, drug resistance, hematopoietic progenitor cell gene transfer, cancer stem cells, cellular therapies, homologous recombination, ribozyme technology, antisense technology, tumor immunotherapy and tumor suppressors, translational research, cancer therapy, gene delivery systems (viral and non-viral), anti-gene therapy (antisense, siRNA & ribozymes), apoptosis; mechanisms and therapies, vaccine development, immunology and immunotherapy, DNA synthesis and repair.
Cancer Gene Therapy publishes the results of laboratory investigations, preclinical studies, and clinical trials in the field of gene transfer/gene therapy and cellular therapies as applied to cancer research. Types of articles published include original research articles; case reports; brief communications; review articles in the main fields of drug resistance/sensitivity, gene therapy, cellular therapy, tumor suppressor and anti-oncogene therapy, cytokine/tumor immunotherapy, etc.; industry perspectives; and letters to the editor.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


