鉴定和验证新型前列腺癌免疫相关预后模型
IF 1.5
4区 医学
Q4 GENETICS & HEREDITY
引用次数: 0
摘要
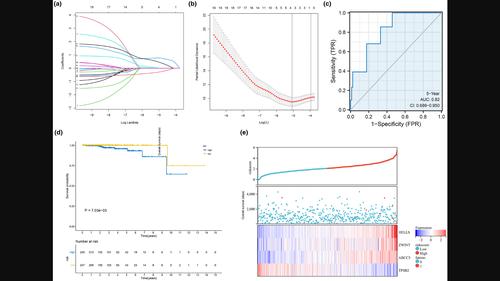
背景肿瘤耐药是致癌转化的一个标志性特征,对肿瘤的进展和转移至关重要。方法 我们从 TCGA 数据库中收集了 495 例 PCa 患者的基因表达谱、单核苷酸多态性突变和拷贝数变异(CNV)数据,并从 MSKCC 数据集中收集了 140 例 PCa 样本。我们提取了434个anoikis相关基因,并利用无监督共识聚类分析确定了分子亚型。对亚型的免疫浸润、分子功能和基因组改变进行了评估。利用 Cox 回归分析建立了风险特征,并通过 MSKCC 数据集进行了验证。我们还确定了针对高危人群的潜在药物。与 C2 相比,C1 表现出更高的 CNV 扩增水平、免疫评分、基质评分、非整倍体评分、同源重组缺陷、瘤内异质性、单核苷酸变异新抗原和肿瘤突变负荷。C2 的生存率更高,γ δ T 细胞和活化 B 细胞浸润水平更高。由四个基因(HELLS、ZWINT、ABCC5 和 TPSB2)组成的风险特征(曲线下面积 = 0.780)被发现是影响 PCa 患者总生存期的独立预后因素。结论本研究中开发的anoikis相关预后模型可作为临床决策的有用工具。这项研究可能会为anoikis相关PCa的治疗提供一个新的视角。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Identification and validation of a novel anoikis‐related prognostic model for prostate cancer
BackgroundAnoikis resistance is a hallmark characteristic of oncogenic transformation, which is crucial for tumor progression and metastasis. The aim of this study was to identify and validate a novel anoikis‐related prognostic model for prostate cancer (PCa).MethodsWe collected a gene expression profile, single nucleotide polymorphism mutation and copy number variation (CNV) data of 495 PCa patients from the TCGA database and 140 PCa samples from the MSKCC dataset. We extracted 434 anoikis‐related genes and unsupervised consensus cluster analysis was used to identify molecular subtypes. The immune infiltration, molecular function, and genome alteration of subtypes were evaluated. A risk signature was developed using Cox regression analysis and validated with the MSKCC dataset. We also identify potential drugs for high‐risk group patients.ResultsTwo subtypes were identified. C1 exhibited a higher level of CNV amplification, immune score, stromal score, aneuploidy score, homologous recombination deficiency, intratumor heterogeneity, single‐nucleotide variant neoantigens, and tumor mutational burden compared to C2. C2 showed a better survival outcome and had a high level of gamma delta T cell and activated B cell infiltration. The risk signature consisting of four genes (HELLS, ZWINT, ABCC5, and TPSB2) was developed (area under the curve = 0.780) and was found to be an independent prognostic factor for overall survival in PCa patients. Four CTRP‐derived and four PRISM‐derived compounds were identified for high‐risk patients.ConclusionsThe anoikis‐related prognostic model developed in this study could be a useful tool for clinical decision‐making. This study may provide a new perspective for the treatment of anoikis‐related PCa.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Genetics & Genomic Medicine
Biochemistry, Genetics and Molecular Biology-Genetics
CiteScore
4.20
自引率
0.00%
发文量
241
审稿时长
14 weeks
期刊介绍:
Molecular Genetics & Genomic Medicine is a peer-reviewed journal for rapid dissemination of quality research related to the dynamically developing areas of human, molecular and medical genetics. The journal publishes original research articles covering findings in phenotypic, molecular, biological, and genomic aspects of genomic variation, inherited disorders and birth defects. The broad publishing spectrum of Molecular Genetics & Genomic Medicine includes rare and common disorders from diagnosis to treatment. Examples of appropriate articles include reports of novel disease genes, functional studies of genetic variants, in-depth genotype-phenotype studies, genomic analysis of inherited disorders, molecular diagnostic methods, medical bioinformatics, ethical, legal, and social implications (ELSI), and approaches to clinical diagnosis. Molecular Genetics & Genomic Medicine provides a scientific home for next generation sequencing studies of rare and common disorders, which will make research in this fascinating area easily and rapidly accessible to the scientific community. This will serve as the basis for translating next generation sequencing studies into individualized diagnostics and therapeutics, for day-to-day medical care.
Molecular Genetics & Genomic Medicine publishes original research articles, reviews, and research methods papers, along with invited editorials and commentaries. Original research papers must report well-conducted research with conclusions supported by the data presented.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


