发现 TrkB 受体结构域 5 的口袋网络--寻找新配体的潜在新目标
IF 3.1
4区 医学
Q3 CHEMISTRY, MEDICINAL
引用次数: 0
摘要
神经营养素酪氨酸激酶受体(TrkB)在阿尔茨海默病、帕金森病、亨廷顿病等多种神经退行性疾病的发病机制中发挥着重要作用,这一点已得到充分描述。这并不奇怪,因为在生理条件下,一旦被脑源性神经营养因子(BDNF)和神经营养素-4/5(NT-4/5)激活,TrkB 受体就会促进神经元的存活、分化和突触功能。考虑到 TrkB 受体的天然配体是大型蛋白质,发现能够模拟其效应的小分子是一项挑战。尽管与 BDNF 或 NT-4/5 相互作用的受体表面已被知晓,但一直存在的问题是,哪个口袋和相互作用负责激活它。为了回答这个具有挑战性的问题,我们利用分子动力学(MD)模拟和 Pocketron 算法,首次发现了存在于受体相互作用结构域(d5)中的口袋网络,并对其进行了描述,了解了它们是如何相互沟通的。这一新发现为我们提供了受体上潜在的新区域,我们可以将其作为目标,并在开发新配体时采用基于结构的药物设计方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
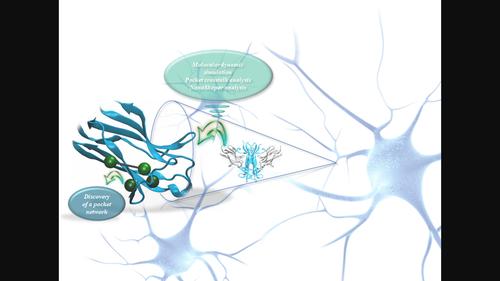
Discovery of a pocket network on the domain 5 of the TrkB receptor – A potential new target in the quest for the new ligands
The important role that the neurotrophin tyrosine kinase receptor ‐ TrkB has in the pathogenesis of several neurodegenerative conditions such are Alzheimer's disease, Parkinson's disease, Huntington's disease, has been well described. This shouldn't be a surprise, since in the physiological conditions, once activated by brain‐derived neurotrophic factor (BDNF) and neurotrophin‐4/5 (NT‐4/5), the TrkB receptor promotes neuronal survival, differentiation and synaptic function. Considering that the natural ligands for TrkB receptor are large proteins, it is a challenge to discover small molecule capable to mimic their effects.Even though, the surface of receptor that is interacting with BDNF or NT‐4/5 is known, there was always a question which pocket and interaction is responsible for activation of it. In order to answer this challenging question, we have used molecular dynamic (MD) simulations and Pocketron algorithm which enabled us to detect, for the first time, a pocket network existing in the interacting domain (d5) of the receptor; to describe them and to see how they are communicating with each other. This new discovery gave us potential new areas on receptor that can be targeted and used for structure‐based drug design approach in the development of the new ligands.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Informatics
CHEMISTRY, MEDICINAL-MATHEMATICAL & COMPUTATIONAL BIOLOGY
CiteScore
7.30
自引率
2.80%
发文量
70
审稿时长
3 months
期刊介绍:
Molecular Informatics is a peer-reviewed, international forum for publication of high-quality, interdisciplinary research on all molecular aspects of bio/cheminformatics and computer-assisted molecular design. Molecular Informatics succeeded QSAR & Combinatorial Science in 2010.
Molecular Informatics presents methodological innovations that will lead to a deeper understanding of ligand-receptor interactions, macromolecular complexes, molecular networks, design concepts and processes that demonstrate how ideas and design concepts lead to molecules with a desired structure or function, preferably including experimental validation.
The journal''s scope includes but is not limited to the fields of drug discovery and chemical biology, protein and nucleic acid engineering and design, the design of nanomolecular structures, strategies for modeling of macromolecular assemblies, molecular networks and systems, pharmaco- and chemogenomics, computer-assisted screening strategies, as well as novel technologies for the de novo design of biologically active molecules. As a unique feature Molecular Informatics publishes so-called "Methods Corner" review-type articles which feature important technological concepts and advances within the scope of the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


