V. V. Sharutin
求助PDF
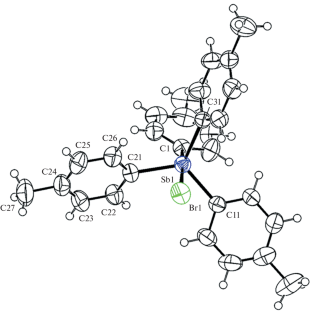
{"title":"四(对甲苯)锑 p-Tol4SbX (X = Br, OC(O)Ph ∙ PhH, OSO2C6H2Me3-2,4,6) 的结构特征衍生物","authors":"V. V. Sharutin","doi":"10.1134/s2634827624600026","DOIUrl":null,"url":null,"abstract":"<h3 data-test=\"abstract-sub-heading\">Abstract</h3><p>The structure of the tetra(<i>p</i>-tolyl)antimony compounds <i>p</i>-Tol<sub>4</sub>SbX (X = Br (<b>1</b>), OC(O)Ph ∙ PhH (<b>2</b>), OSO<sub>2</sub>C<sub>6</sub>Me<sub>3</sub>-2,4,6 (<b>3</b>)) is determined by X-ray diffraction analysis (XRD). According to the XRD data, the antimony atoms in complexes <b>1</b>–<b>3</b> exhibit a distorted trigonal bipyramidal coordination with three aryl ligands in the equatorial plane, and the axial angles CSbX are 174.75(8)°, 175.13(9)°, and 174.51(6)°, respectively.</p><p>The XRD data for compound <b>1</b> are as follows: C<sub>28</sub>H<sub>28</sub>BrSb, <i>M</i> = 566.16; monoclinic system, sp. gr. <i>P</i>2<sub>1</sub>/<i>n</i>; cell parameters: <i>a</i> = 9.868(6) Å, <i>b</i> = 23.312(11) Å, <i>c =</i> 12.106(6) Å; β = 113.15(2)°, <i>V</i> = 2561(2) Å<sup>3</sup>, <i>Z</i> = 4; ρ(calc.) = 1.469 g/cm<sup>3</sup>; μ = 2.649 mm<sup>–1</sup>; <i>F</i>(000) = 1128.0; 2θ data acquisition region: 6.4°–56.76°; –13 ≤ <i>h</i> ≤ 13, –31 ≤ <i>k</i> ≤ 31, –16 ≤ <i>l</i> ≤ 16; total reflections 42 998; independent reflections 6359 (<i>R</i><sub>int</sub> = 0.0346); GOOF = 1.080; <i>R</i> factor 0.0325. For compound <b>2</b>: C<sub>41</sub>H<sub>39</sub>O<sub>2</sub>Sb, <i>M</i> = 685.47; monoclinic system, sp. gr. <i>С</i>2/<i>с</i>; cell parameters: <i>a</i> = 28.186(13) Å, <i>b</i> = 15.116(6) Å, <i>c =</i> 17.629(8) Å; β = 91.73(2)°, <i>V</i> = 7507(6) Å<sup>3</sup>, <i>Z</i> = 8; ρ(calc.) = 1.213 g/cm<sup>3</sup>; μ = 0.765 mm<sup>–1</sup>; <i>F</i>(000) = 2816,0; 2θ data acquisition region: 6.572°–56.996°; –37 ≤ <i>h</i> ≤ 37, –20 ≤ <i>k</i> ≤ 20, –23 ≤ <i>l</i> ≤ 23; total reflections 116 806; independent reflections 9489 (<i>R</i><sub>int</sub> = 0.0492); GOOF = 1.102; <i>R</i> factor 0.0363. For compound <b>3</b>: C<sub>37</sub>H<sub>39</sub>O<sub>3</sub>SSb, <i>M</i> = 685.49; monoclinic system, sp. gr. <i>P</i>2<sub>1</sub>/<i>n</i>; cell parametrs: <i>a</i> = 12.172(4) Å, <i>b</i> = 18.802(5) Å, <i>c =</i> 15.433(6) Å; β = 108.744(12)°, <i>V</i> = 3345(2) Å<sup>3</sup>, <i>Z</i> = 4; ρ(calc.) = 1.361 g/cm<sup>3</sup>; μ = 0.921 mm<sup>–1</sup>; <i>F</i>(000) = 1408.0; 2θ data acquisition region: 5.96°–63.02°; –16 ≤ <i>h</i> ≤ 17, –27 ≤ <i>k</i> ≤ 27, ‒22 ≤ <i>l</i> ≤ 21; total reflections 138 835; independent reflections 11 081 (<i>R</i><sub>int</sub> = 0.0373); GOOF = 1.045; <i>R</i> factor 0.0304. Complete tables of atomic coordinates, bond lengths, and bond angles for compounds <b>1</b>–<b>3</b> have been deposited to the Cambridge Structural Database (CSD) with the following deposition numbers: 2182608, 2149953, and 2171918. For access to the data, please contact deposit@ccdc.cam.ac.uk or visit http://www.ccdc.cam.ac.uk.</p>","PeriodicalId":21086,"journal":{"name":"Reviews and Advances in Chemistry","volume":"51 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-03-20","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"","citationCount":"0","resultStr":"{\"title\":\"Structural Features of Tetra(p-tolyl)antimony p-Tol4SbX (X = Br, OC(O)Ph ∙ PhH, OSO2C6H2Me3-2,4,6) Derivatives\",\"authors\":\"V. V. Sharutin\",\"doi\":\"10.1134/s2634827624600026\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<h3 data-test=\\\"abstract-sub-heading\\\">Abstract</h3><p>The structure of the tetra(<i>p</i>-tolyl)antimony compounds <i>p</i>-Tol<sub>4</sub>SbX (X = Br (<b>1</b>), OC(O)Ph ∙ PhH (<b>2</b>), OSO<sub>2</sub>C<sub>6</sub>Me<sub>3</sub>-2,4,6 (<b>3</b>)) is determined by X-ray diffraction analysis (XRD). According to the XRD data, the antimony atoms in complexes <b>1</b>–<b>3</b> exhibit a distorted trigonal bipyramidal coordination with three aryl ligands in the equatorial plane, and the axial angles CSbX are 174.75(8)°, 175.13(9)°, and 174.51(6)°, respectively.</p><p>The XRD data for compound <b>1</b> are as follows: C<sub>28</sub>H<sub>28</sub>BrSb, <i>M</i> = 566.16; monoclinic system, sp. gr. <i>P</i>2<sub>1</sub>/<i>n</i>; cell parameters: <i>a</i> = 9.868(6) Å, <i>b</i> = 23.312(11) Å, <i>c =</i> 12.106(6) Å; β = 113.15(2)°, <i>V</i> = 2561(2) Å<sup>3</sup>, <i>Z</i> = 4; ρ(calc.) = 1.469 g/cm<sup>3</sup>; μ = 2.649 mm<sup>–1</sup>; <i>F</i>(000) = 1128.0; 2θ data acquisition region: 6.4°–56.76°; –13 ≤ <i>h</i> ≤ 13, –31 ≤ <i>k</i> ≤ 31, –16 ≤ <i>l</i> ≤ 16; total reflections 42 998; independent reflections 6359 (<i>R</i><sub>int</sub> = 0.0346); GOOF = 1.080; <i>R</i> factor 0.0325. For compound <b>2</b>: C<sub>41</sub>H<sub>39</sub>O<sub>2</sub>Sb, <i>M</i> = 685.47; monoclinic system, sp. gr. <i>С</i>2/<i>с</i>; cell parameters: <i>a</i> = 28.186(13) Å, <i>b</i> = 15.116(6) Å, <i>c =</i> 17.629(8) Å; β = 91.73(2)°, <i>V</i> = 7507(6) Å<sup>3</sup>, <i>Z</i> = 8; ρ(calc.) = 1.213 g/cm<sup>3</sup>; μ = 0.765 mm<sup>–1</sup>; <i>F</i>(000) = 2816,0; 2θ data acquisition region: 6.572°–56.996°; –37 ≤ <i>h</i> ≤ 37, –20 ≤ <i>k</i> ≤ 20, –23 ≤ <i>l</i> ≤ 23; total reflections 116 806; independent reflections 9489 (<i>R</i><sub>int</sub> = 0.0492); GOOF = 1.102; <i>R</i> factor 0.0363. For compound <b>3</b>: C<sub>37</sub>H<sub>39</sub>O<sub>3</sub>SSb, <i>M</i> = 685.49; monoclinic system, sp. gr. <i>P</i>2<sub>1</sub>/<i>n</i>; cell parametrs: <i>a</i> = 12.172(4) Å, <i>b</i> = 18.802(5) Å, <i>c =</i> 15.433(6) Å; β = 108.744(12)°, <i>V</i> = 3345(2) Å<sup>3</sup>, <i>Z</i> = 4; ρ(calc.) = 1.361 g/cm<sup>3</sup>; μ = 0.921 mm<sup>–1</sup>; <i>F</i>(000) = 1408.0; 2θ data acquisition region: 5.96°–63.02°; –16 ≤ <i>h</i> ≤ 17, –27 ≤ <i>k</i> ≤ 27, ‒22 ≤ <i>l</i> ≤ 21; total reflections 138 835; independent reflections 11 081 (<i>R</i><sub>int</sub> = 0.0373); GOOF = 1.045; <i>R</i> factor 0.0304. Complete tables of atomic coordinates, bond lengths, and bond angles for compounds <b>1</b>–<b>3</b> have been deposited to the Cambridge Structural Database (CSD) with the following deposition numbers: 2182608, 2149953, and 2171918. For access to the data, please contact deposit@ccdc.cam.ac.uk or visit http://www.ccdc.cam.ac.uk.</p>\",\"PeriodicalId\":21086,\"journal\":{\"name\":\"Reviews and Advances in Chemistry\",\"volume\":\"51 1\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2024-03-20\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Reviews and Advances in Chemistry\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://doi.org/10.1134/s2634827624600026\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"\",\"JCRName\":\"\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Reviews and Advances in Chemistry","FirstCategoryId":"1085","ListUrlMain":"https://doi.org/10.1134/s2634827624600026","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
引用
批量引用
Structural Features of Tetra(p-tolyl)antimony p-Tol4SbX (X = Br, OC(O)Ph ∙ PhH, OSO2C6H2Me3-2,4,6) Derivatives
Abstract The structure of the tetra(p -tolyl)antimony compounds p -Tol4 SbX (X = Br (1 ), OC(O)Ph ∙ PhH (2 ), OSO2 C6 Me3 -2,4,6 (3 )) is determined by X-ray diffraction analysis (XRD). According to the XRD data, the antimony atoms in complexes 1 –3 exhibit a distorted trigonal bipyramidal coordination with three aryl ligands in the equatorial plane, and the axial angles CSbX are 174.75(8)°, 175.13(9)°, and 174.51(6)°, respectively.
The XRD data for compound 1 are as follows: C28 H28 BrSb, M = 566.16; monoclinic system, sp. gr. P 21 /n ; cell parameters: a = 9.868(6) Å, b = 23.312(11) Å, c = 12.106(6) Å; β = 113.15(2)°, V = 2561(2) Å3 , Z = 4; ρ(calc.) = 1.469 g/cm3 ; μ = 2.649 mm–1 ; F (000) = 1128.0; 2θ data acquisition region: 6.4°–56.76°; –13 ≤ h ≤ 13, –31 ≤ k ≤ 31, –16 ≤ l ≤ 16; total reflections 42 998; independent reflections 6359 (R int = 0.0346); GOOF = 1.080; R factor 0.0325. For compound 2 : C41 H39 O2 Sb, M = 685.47; monoclinic system, sp. gr. С 2/с ; cell parameters: a = 28.186(13) Å, b = 15.116(6) Å, c = 17.629(8) Å; β = 91.73(2)°, V = 7507(6) Å3 , Z = 8; ρ(calc.) = 1.213 g/cm3 ; μ = 0.765 mm–1 ; F (000) = 2816,0; 2θ data acquisition region: 6.572°–56.996°; –37 ≤ h ≤ 37, –20 ≤ k ≤ 20, –23 ≤ l ≤ 23; total reflections 116 806; independent reflections 9489 (R int = 0.0492); GOOF = 1.102; R factor 0.0363. For compound 3 : C37 H39 O3 SSb, M = 685.49; monoclinic system, sp. gr. P 21 /n ; cell parametrs: a = 12.172(4) Å, b = 18.802(5) Å, c = 15.433(6) Å; β = 108.744(12)°, V = 3345(2) Å3 , Z = 4; ρ(calc.) = 1.361 g/cm3 ; μ = 0.921 mm–1 ; F (000) = 1408.0; 2θ data acquisition region: 5.96°–63.02°; –16 ≤ h ≤ 17, –27 ≤ k ≤ 27, ‒22 ≤ l ≤ 21; total reflections 138 835; independent reflections 11 081 (R int = 0.0373); GOOF = 1.045; R factor 0.0304. Complete tables of atomic coordinates, bond lengths, and bond angles for compounds 1 –3 have been deposited to the Cambridge Structural Database (CSD) with the following deposition numbers: 2182608, 2149953, and 2171918. For access to the data, please contact deposit@ccdc.cam.ac.uk or visit http://www.ccdc.cam.ac.uk.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


