单细胞和空间转录组 RNA-seq 的整合分析揭示了 ccRCC 的肿瘤异质性、治疗靶点和预后亚型。
IF 4.8
3区 医学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
透明细胞肾细胞癌(ccRCC)是最常见的RCC类型;然而,ccRCC的瘤内异质性仍不清楚。我们首先利用基于单细胞和空间转录组 RNA 序列数据的生物信息学分析确定了每个细胞群的标记和生物学特征。我们发现,3p染色体上的基因拷贝数丢失和5q染色体上的基因扩增是ccRCC细胞的共同特征。同时,与缺氧反应和新陈代谢相关的NNMT和HILPDA是ccRCC的潜在治疗靶点。此外,与免疫抑制密切相关的CD8+衰竭T细胞(LAG3+ HAVCR2+)、CD8+增殖T细胞(STMN+)和M2样巨噬细胞(CD68+ CD163+ APOC1+)在ccRCC的发生和发展中发挥着重要作用。全外显子组测序、细胞系和异种移植实验以及免疫荧光染色进一步验证了这些结果。最后,我们利用无监督聚类分析将ccRCC患者分为三个亚型,并利用梯度提升算法(eXtreme Gradient Boosting algorithm)生成了一个分类器来再现这些亚型。我们的分类器可以帮助临床医生评估 ccRCC 的预后并设计个性化的治疗策略。总之,我们的工作为理解肿瘤异质性提供了一个新的视角,将有助于ccRCC抗肿瘤治疗策略的设计。本文章由计算机程序翻译,如有差异,请以英文原文为准。
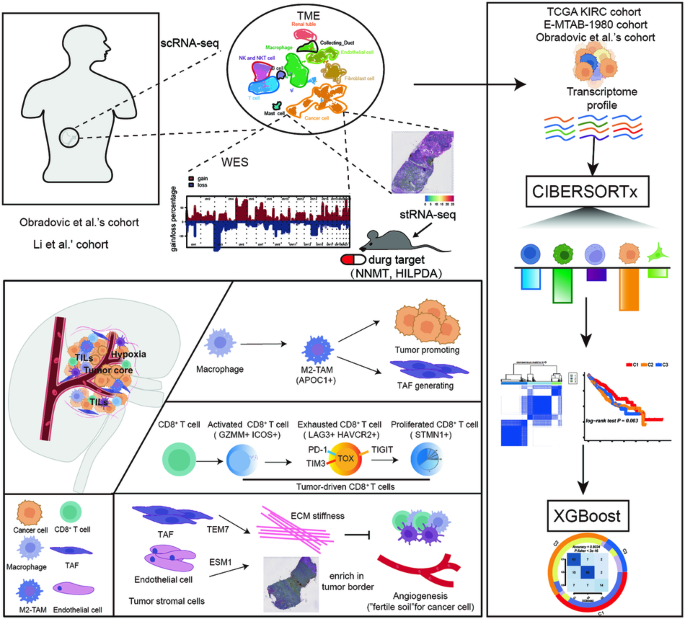
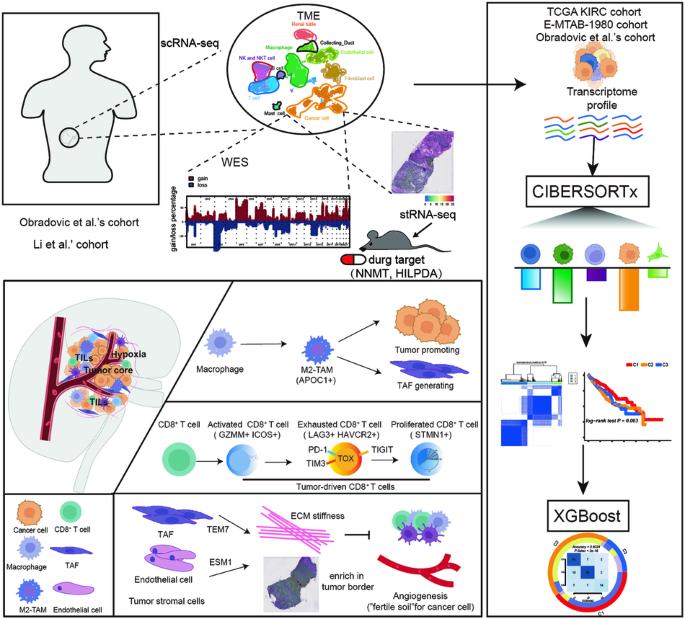
The integrate profiling of single-cell and spatial transcriptome RNA-seq reveals tumor heterogeneity, therapeutic targets, and prognostic subtypes in ccRCC
Clear-cell renal cell carcinoma (ccRCC) is the most common type of RCC; however, the intratumoral heterogeneity in ccRCC remains unclear. We first identified markers and biological features of each cell cluster using bioinformatics analysis based on single-cell and spatial transcriptome RNA-sequencing data. We found that gene copy number loss on chromosome 3p and amplification on chromosome 5q were common features in ccRCC cells. Meanwhile, NNMT and HILPDA, which are associated with the response to hypoxia and metabolism, are potential therapeutic targets for ccRCC. In addition, CD8+ exhausted T cells (LAG3+ HAVCR2+), CD8+ proliferated T cells (STMN+), and M2-like macrophages (CD68+ CD163+ APOC1+), which are closely associated with immunosuppression, played vital roles in ccRCC occurrence and development. These results were further verified by whole exome sequencing, cell line and xenograft experiments, and immunofluorescence staining. Finally, we divide patients with ccRCC into three subtypes using unsupervised cluster analysis. and generated a classifier to reproduce these subtypes using the eXtreme Gradient Boosting algorithm. Our classifier can help clinicians evaluate prognosis and design personalized treatment strategies for ccRCC. In summary, our work provides a new perspective for understanding tumor heterogeneity and will aid in the design of antitumor therapeutic strategies for ccRCC.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cancer gene therapy
医学-生物工程与应用微生物
CiteScore
10.20
自引率
0.00%
发文量
150
审稿时长
4-8 weeks
期刊介绍:
Cancer Gene Therapy is the essential gene and cellular therapy resource for cancer researchers and clinicians, keeping readers up to date with the latest developments in gene and cellular therapies for cancer. The journal publishes original laboratory and clinical research papers, case reports and review articles. Publication topics include RNAi approaches, drug resistance, hematopoietic progenitor cell gene transfer, cancer stem cells, cellular therapies, homologous recombination, ribozyme technology, antisense technology, tumor immunotherapy and tumor suppressors, translational research, cancer therapy, gene delivery systems (viral and non-viral), anti-gene therapy (antisense, siRNA & ribozymes), apoptosis; mechanisms and therapies, vaccine development, immunology and immunotherapy, DNA synthesis and repair.
Cancer Gene Therapy publishes the results of laboratory investigations, preclinical studies, and clinical trials in the field of gene transfer/gene therapy and cellular therapies as applied to cancer research. Types of articles published include original research articles; case reports; brief communications; review articles in the main fields of drug resistance/sensitivity, gene therapy, cellular therapy, tumor suppressor and anti-oncogene therapy, cytokine/tumor immunotherapy, etc.; industry perspectives; and letters to the editor.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


