利用从海绵副渔获标本中获得的eDNA表征南极鱼类组合
IF 4.6
1区 农林科学
Q1 FISHERIES
引用次数: 0
摘要
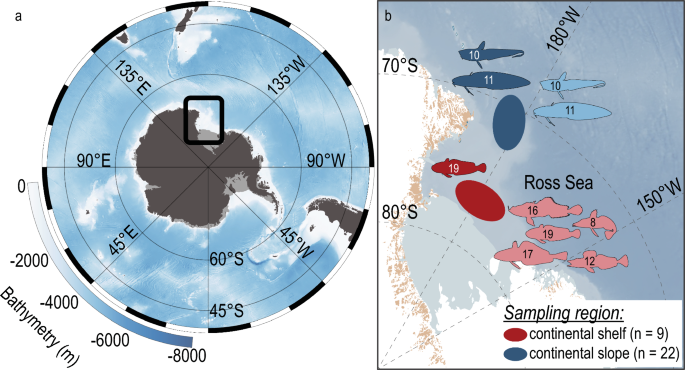
国际保护目标是缓解南大洋生态系统的恶化,并通过多个监测项目评估这些目标的进展。然而,通过目视观察进行持续监测的规模受到该地区偏远和后勤限制的挑战。鉴于南大洋的生态和经济重要性,必须探索更多的生物监测方法。最近,在南大洋渔业中经常捕获和丢弃的海洋海绵被证明可以自然积累环境DNA (eDNA)。在这里,我们比较了来自海绵副渔获标本的鱼类eDNA信号与来自南极洲罗斯海大陆架(523.5-709米)的9个地点和大陆斜坡(887.5-1611.5米)的17个地点的鱼类捕捞记录。我们共记录了20条鱼,其中12条为捕获鱼,18条为eDNA观察,10条为两种方法均检测到。虽然采样位置是数据集中观察到的变化的最大贡献者,但与渔获记录相比,eDNA获得了更高的物种丰富度,并显示出明显不同的物种组成。总体而言,eDNA读取数与鱼类生物量丰度的相关性更强。物种组成在区域尺度上具有相关性,但eDNA信号强度在物种水平上不能预测捕获数量。与捕获记录相比,我们的研究结果突出了在南大洋中通过检测更大比例的鱼类群落来监测海绵eDNA的潜力,从而增加了我们对这一未被充分研究的生态系统的了解,并最终帮助保护工作。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Characterizing Antarctic fish assemblages using eDNA obtained from marine sponge bycatch specimens
Abstract International conservation goals have been set to mitigate Southern Ocean ecosystem deterioration, with multiple monitoring programs evaluating progress towards those goals. The scale of continuous monitoring through visual observations, however, is challenged by the remoteness of the area and logistical constraints. Given the ecological and economic importance of the Southern Ocean, it is imperative that additional biological monitoring approaches are explored. Recently, marine sponges, which are frequently caught and discarded in Southern Ocean fisheries, have been shown to naturally accumulate environmental DNA (eDNA). Here, we compare fish eDNA signals from marine sponge bycatch specimens to fish catch records for nine locations on the continental shelf (523.5–709 m) and 17 from the continental slope (887.5–1611.5 m) within the Ross Sea, Antarctica. We recorded a total of 20 fishes, with 12 fishes reported as catch, 18 observed by eDNA, and ten detected by both methods. While sampling location was the largest contributor to the variation observed in the dataset, eDNA obtained significantly higher species richness and displayed a significantly different species composition compared to fish catch records. Overall, eDNA read count correlated more strongly with fish abundance over biomass. Species composition correlated on a regional scale between methods, however eDNA signal strength was a low predictor of catch numbers at the species level. Our results highlight the potential of sponge eDNA monitoring in the Southern Ocean by detecting a larger fraction of the fish community compared to catch recordings, thereby increasing our knowledge of this understudied ecosystem and, ultimately, aiding conservation efforts.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Reviews in Fish Biology and Fisheries
农林科学-海洋与淡水生物学
CiteScore
10.00
自引率
8.10%
发文量
42
审稿时长
12-24 weeks
期刊介绍:
The subject matter is focused on include evolutionary biology, zoogeography, taxonomy, including biochemical taxonomy and stock identification, genetics and genetic manipulation, physiology, functional morphology, behaviour, ecology, fisheries assessment, development, exploitation and conservation. however, reviews will be published from any field of fish biology where the emphasis is placed on adaptation, function or exploitation in the whole organism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


