高维数据的联邦奇异值分解
IF 2.8
3区 计算机科学
Q2 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
联邦学习(FL)正在成为传统基于云的机器学习的隐私意识替代品。在FL中,敏感数据保留在数据孤岛中,只交换聚合参数。不愿意分享数据的医院和研究机构可以加入联合研究,而不会违反保密规定。除了生物医学数据的极端敏感性外,高维度对联合全基因组关联研究(GWAS)提出了挑战。在本文中,我们提出了一种联邦奇异值分解算法,该算法适用于GWAS的隐私相关和计算需求。值得注意的是,该算法的传输成本与样本数量无关,仅弱依赖于特征数量,因为样本对应的奇异向量永远不会交换,与特征相关的向量仅在固定次数的迭代中传输到聚合器。虽然该算法是由GWAS驱动的,但它一般适用于水平和垂直分区的数据。本文章由计算机程序翻译,如有差异,请以英文原文为准。
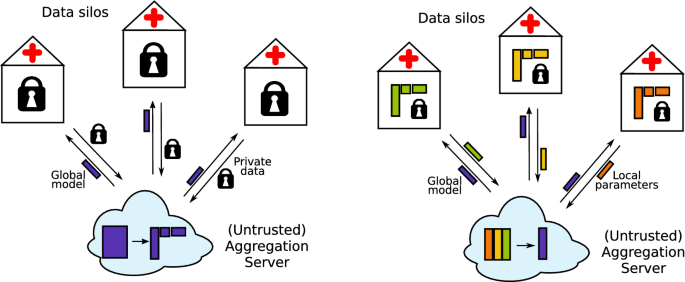
Federated singular value decomposition for high-dimensional data
Abstract Federated learning (FL) is emerging as a privacy-aware alternative to classical cloud-based machine learning. In FL, the sensitive data remains in data silos and only aggregated parameters are exchanged. Hospitals and research institutions which are not willing to share their data can join a federated study without breaching confidentiality. In addition to the extreme sensitivity of biomedical data, the high dimensionality poses a challenge in the context of federated genome-wide association studies (GWAS). In this article, we present a federated singular value decomposition algorithm, suitable for the privacy-related and computational requirements of GWAS. Notably, the algorithm has a transmission cost independent of the number of samples and is only weakly dependent on the number of features, because the singular vectors corresponding to the samples are never exchanged and the vectors associated with the features are only transmitted to an aggregator for a fixed number of iterations. Although motivated by GWAS, the algorithm is generically applicable for both horizontally and vertically partitioned data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Data Mining and Knowledge Discovery
工程技术-计算机:人工智能
CiteScore
10.40
自引率
4.20%
发文量
68
审稿时长
10 months
期刊介绍:
Advances in data gathering, storage, and distribution have created a need for computational tools and techniques to aid in data analysis. Data Mining and Knowledge Discovery in Databases (KDD) is a rapidly growing area of research and application that builds on techniques and theories from many fields, including statistics, databases, pattern recognition and learning, data visualization, uncertainty modelling, data warehousing and OLAP, optimization, and high performance computing.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


