inStrain 从元基因组数据中剖析种群微多样性,灵敏检测共有微生物菌株
IF 41.7
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 170
摘要
同一物种共存的微生物细胞往往会出现遗传变异,而遗传变异会影响从营养偏好到致病性等各种表型。我们在这里介绍 inStrain,它是一种利用元基因组配对读数来描述整个基因组种群内遗传多样性(微多样性)的程序,并以微多样性感知的方式对微生物种群进行比较,从而大大提高了与现有方法进行基因组比较的准确性。我们使用 inStrain 分析了来自早产新生儿的 1000 个粪便元基因组,发现同胞兄弟姐妹共享的菌株明显多于无血缘关系的婴儿,尽管同卵双胞胎共享的菌株并不比异卵双胞胎兄弟姐妹多。剖腹产婴儿携带的克雷伯氏菌的核苷酸多样性明显高于阴道分娩婴儿,这可能反映了婴儿是从医院而非母体微生物组中获得的。在婴儿个体中显示出多样性的基因组位点包括在其他婴儿之间发现的变体,这可能反映出接种来自不同的医院相关来源。元基因组分析工具可识别种群内和种群间的微生物菌株。本文章由计算机程序翻译,如有差异,请以英文原文为准。
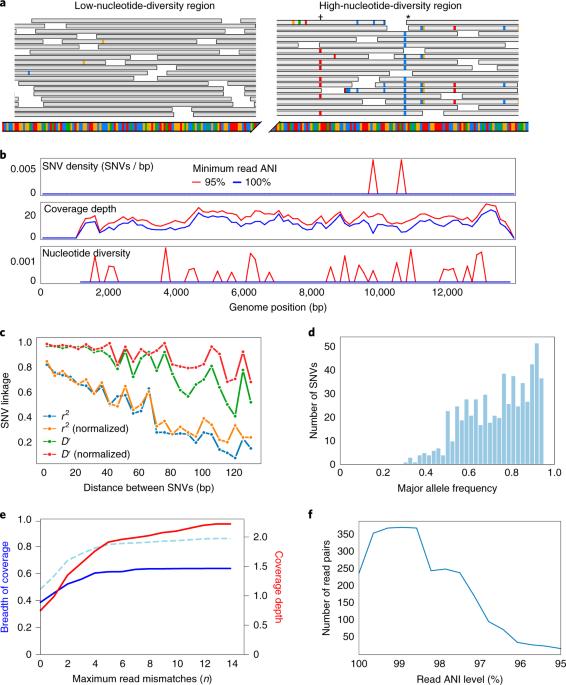
inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains
Coexisting microbial cells of the same species often exhibit genetic variation that can affect phenotypes ranging from nutrient preference to pathogenicity. Here we present inStrain, a program that uses metagenomic paired reads to profile intra-population genetic diversity (microdiversity) across whole genomes and compares microbial populations in a microdiversity-aware manner, greatly increasing the accuracy of genomic comparisons when benchmarked against existing methods. We use inStrain to profile >1,000 fecal metagenomes from newborn premature infants and find that siblings share significantly more strains than unrelated infants, although identical twins share no more strains than fraternal siblings. Infants born by cesarean section harbor Klebsiella with significantly higher nucleotide diversity than infants delivered vaginally, potentially reflecting acquisition from hospital rather than maternal microbiomes. Genomic loci that show diversity in individual infants include variants found between other infants, possibly reflecting inoculation from diverse hospital-associated sources. inStrain can be applied to any metagenomic dataset for microdiversity analysis and rigorous strain comparison. A metagenome analysis tool identifies microbial strains within and between populations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature biotechnology
工程技术-生物工程与应用微生物
CiteScore
63.00
自引率
1.70%
发文量
382
审稿时长
3 months
期刊介绍:
Nature Biotechnology is a monthly journal that focuses on the science and business of biotechnology. It covers a wide range of topics including technology/methodology advancements in the biological, biomedical, agricultural, and environmental sciences. The journal also explores the commercial, political, ethical, legal, and societal aspects of this research.
The journal serves researchers by providing peer-reviewed research papers in the field of biotechnology. It also serves the business community by delivering news about research developments. This approach ensures that both the scientific and business communities are well-informed and able to stay up-to-date on the latest advancements and opportunities in the field.
Some key areas of interest in which the journal actively seeks research papers include molecular engineering of nucleic acids and proteins, molecular therapy, large-scale biology, computational biology, regenerative medicine, imaging technology, analytical biotechnology, applied immunology, food and agricultural biotechnology, and environmental biotechnology.
In summary, Nature Biotechnology is a comprehensive journal that covers both the scientific and business aspects of biotechnology. It strives to provide researchers with valuable research papers and news while also delivering important scientific advancements to the business community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


