在单细胞中直接测量工程癌症突变及其转录表型。
IF 33.1
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
基因组测序研究发现了多种肿瘤类型中的大量癌症突变,但确定这些突变的表型结果仍是一项挑战。在这里,我们开发了一种名为TISCC-seq的高通量、多重单细胞技术,利用CRISPR碱基编辑器在细胞中设计预先指定的突变,在单个细胞中直接划分其基因型,并确定每个突变的转录表型。对目标基因的转录本进行长读程测序可确定工程突变,同时通过短读程测序分析同一组细胞的转录组概况。通过整合,我们以单细胞分辨率确定了突变的基因型和表达表型。我们利用细胞系,设计并评估了 >100 个 TP53 基因突变对基因表达的影响。根据单细胞基因表达,我们将突变归类为具有重要功能的表型。本文章由计算机程序翻译,如有差异,请以英文原文为准。
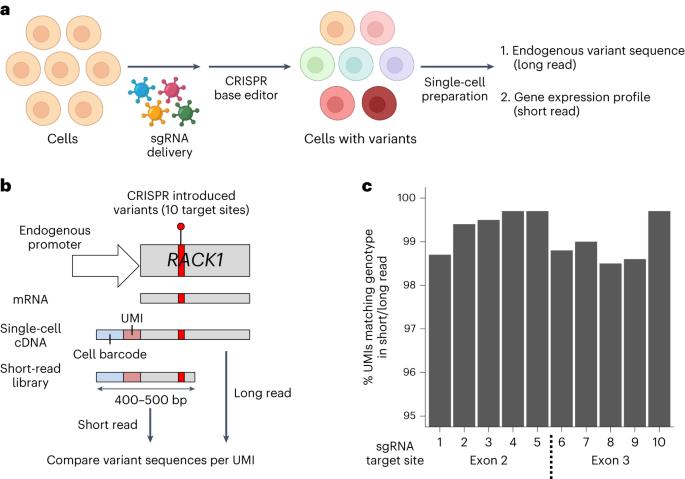
Direct measurement of engineered cancer mutations and their transcriptional phenotypes in single cells
Genome sequencing studies have identified numerous cancer mutations across a wide spectrum of tumor types, but determining the phenotypic consequence of these mutations remains a challenge. Here, we developed a high-throughput, multiplexed single-cell technology called TISCC-seq to engineer predesignated mutations in cells using CRISPR base editors, directly delineate their genotype among individual cells and determine each mutation’s transcriptional phenotype. Long-read sequencing of the target gene’s transcript identifies the engineered mutations, and the transcriptome profile from the same set of cells is simultaneously analyzed by short-read sequencing. Through integration, we determine the mutations’ genotype and expression phenotype at single-cell resolution. Using cell lines, we engineer and evaluate the impact of >100 TP53 mutations on gene expression. Based on the single-cell gene expression, we classify the mutations as having a functionally significant phenotype. Engineered cancer mutations are linked with phenotypes in a multiplexed single-cell technology.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature biotechnology
工程技术-生物工程与应用微生物
CiteScore
63.00
自引率
1.70%
发文量
382
审稿时长
3 months
期刊介绍:
Nature Biotechnology is a monthly journal that focuses on the science and business of biotechnology. It covers a wide range of topics including technology/methodology advancements in the biological, biomedical, agricultural, and environmental sciences. The journal also explores the commercial, political, ethical, legal, and societal aspects of this research.
The journal serves researchers by providing peer-reviewed research papers in the field of biotechnology. It also serves the business community by delivering news about research developments. This approach ensures that both the scientific and business communities are well-informed and able to stay up-to-date on the latest advancements and opportunities in the field.
Some key areas of interest in which the journal actively seeks research papers include molecular engineering of nucleic acids and proteins, molecular therapy, large-scale biology, computational biology, regenerative medicine, imaging technology, analytical biotechnology, applied immunology, food and agricultural biotechnology, and environmental biotechnology.
In summary, Nature Biotechnology is a comprehensive journal that covers both the scientific and business aspects of biotechnology. It strives to provide researchers with valuable research papers and news while also delivering important scientific advancements to the business community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


