Investigating genetic variants for treatment response to selective serotonin reuptake inhibitors in syndromal factors and side effects among patients with depression in Taiwanese Han population
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 1
Abstract
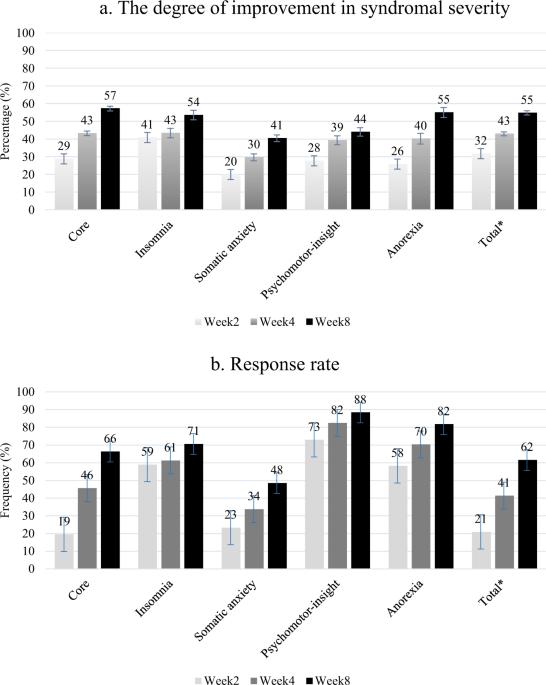
Major depressive disorder (MDD) is associated with high heterogeneity in clinical presentation. In addition, response to treatment with selective serotonin reuptake inhibitors (SSRIs) varies considerably among patients. Therefore, identifying genetic variants that may contribute to SSRI treatment responses in MDD is essential. In this study, we analyzed the syndromal factor structures of the Hamilton Depression Rating Scale in 479 patients with MDD by using exploratory factor analysis. All patients were followed up biweekly for 8 weeks. Treatment response was defined for all syndromal factors and total scores. In addition, a genome-wide association study was performed to investigate the treatment outcomes at week 4 and repeatedly assess all visits during follow-up by using mixed models adjusted for age, gender, and population substructure. Moreover, the role of genetic variants in suicidal and sexual side effects was explored, and five syndromal factors for depression were derived: core, insomnia, somatic anxiety, psychomotor-insight, and anorexia. Subsequently, several known genes were mapped to suggestive signals for treatment outcomes, including single-nucleotide polymorphisms (SNPs) in PRF1, UTP20, MGAM, and ENSG00000286536 for psychomotor-insight and in C4orf51 for anorexia. In total, 33 independent SNPs for treatment responses were tested in a mixed model, 12 of which demonstrated a p value <0.05. The most significant SNP was rs2182717 in the ENSR00000803469 gene located on chromosome 6 for the core syndromal factor (β = −0.638, p = 1.8 × 10−4) in terms of symptom improvement over time. Patients with a GG or GA genotype with the rs2182717 SNP also exhibited a treatment response (β = 0.089, p = 2.0 × 10−6) at week 4. Moreover, rs1836075352 was associated with sexual side effects (p = 3.2 × 10−8). Pathway and network analyses using the identified SNPs revealed potential biological functions involved in treatment response, such as neurodevelopment-related functions and immune processes. In conclusion, we identified loci that may affect the clinical response to treatment with antidepressants in the context of empirically defined depressive syndromal factors and side effects among the Taiwanese Han population, thus providing novel biological targets for further investigation.
调查台湾汉族抑郁症患者对选择性血清素再摄取抑制剂的治疗反应在综合因素和副作用方面的遗传变异
重度抑郁障碍(MDD)的临床表现具有高度异质性。此外,不同患者对选择性血清素再摄取抑制剂(SSRIs)治疗的反应也有很大差异。因此,确定可能会影响 MDD 患者对 SSRI 治疗反应的遗传变异至关重要。在这项研究中,我们采用探索性因子分析方法分析了 479 名 MDD 患者的汉密尔顿抑郁量表(Hamilton Depression Rating Scale)的综合因子结构。所有患者均接受了为期 8 周的双周随访。所有综合因子和总分均定义为治疗反应。此外,还进行了一项全基因组关联研究,通过年龄、性别和人群亚结构调整后的混合模型,调查第 4 周的治疗结果,并在随访期间反复评估所有就诊情况。此外,还探讨了基因变异在自杀和性副作用中的作用,并得出了五个抑郁症综合因素:核心、失眠、躯体焦虑、精神运动-视力和厌食。随后,几个已知基因被映射到治疗结果的提示性信号上,包括PRF1、UTP20、MGAM和ENSG00000286536中的单核苷酸多态性(SNPs)(针对精神运动视力)和C4orf51中的单核苷酸多态性(SNPs)(针对厌食症)。在混合模型中,共有 33 个独立的 SNP 与治疗反应相关,其中 12 个 SNP 的 p 值为 0.05。最显著的 SNP 是位于 6 号染色体上的 ENSR00000803469 基因中的 rs2182717,该 SNP 与核心综合征因子有关(β = -0.638,p = 1.8 × 10-4)。在第 4 周时,rs2182717 SNP 基因型为 GG 或 GA 的患者也表现出治疗反应(β = 0.089,p = 2.0 × 10-6)。此外,rs1836075352 与性副作用相关(p = 3.2 × 10-8)。利用所鉴定的 SNPs 进行的通路和网络分析揭示了参与治疗反应的潜在生物功能,如神经发育相关功能和免疫过程。总之,我们在台湾汉族人群中根据经验定义的抑郁综合征因素和副作用发现了可能影响抗抑郁药物治疗临床反应的位点,从而为进一步研究提供了新的生物学靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


