Predicting microsatellite instability and key biomarkers in colorectal cancer from H&E-stained images: achieving state-of-the-art predictive performance with fewer data using Swin Transformer
IF 3.4
2区 医学
Q1 PATHOLOGY
引用次数: 3
Abstract
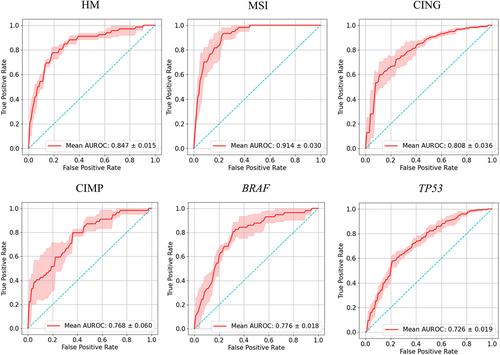
Many artificial intelligence models have been developed to predict clinically relevant biomarkers for colorectal cancer (CRC), including microsatellite instability (MSI). However, existing deep learning networks require large training datasets, which are often hard to obtain. In this study, based on the latest Hierarchical Vision Transformer using Shifted Windows (Swin Transformer [Swin‐T]), we developed an efficient workflow to predict biomarkers in CRC (MSI, hypermutation, chromosomal instability, CpG island methylator phenotype, and BRAF and TP53 mutation) that required relatively small datasets. Our Swin‐T workflow substantially achieved the state‐of‐the‐art (SOTA) predictive performance in an intra‐study cross‐validation experiment on the Cancer Genome Atlas colon and rectal cancer dataset (TCGA‐CRC‐DX). It also demonstrated excellent generalizability in cross‐study external validation and delivered a SOTA area under the receiver operating characteristic curve (AUROC) of 0.90 for MSI, using the Molecular and Cellular Oncology dataset for training (N = 1,065) and the TCGA‐CRC‐DX (N = 462) for testing. A similar performance (AUROC = 0.91) was reported in a recent study, using ~8,000 training samples (ResNet18) on the same testing dataset. Swin‐T was extremely efficient when using small training datasets and exhibited robust predictive performance with 200–500 training samples. Our findings indicate that Swin‐T could be 5–10 times more efficient than existing algorithms for MSI prediction based on ResNet18 and ShuffleNet. Furthermore, the Swin‐T models demonstrated their capability in accurately predicting MSI and BRAF mutation status, which could exclude and therefore reduce samples before subsequent standard testing in a cascading diagnostic workflow, in turn reducing turnaround time and costs.
从h&e染色图像预测结直肠癌的微卫星不稳定性和关键生物标志物:使用Swin Transformer以更少的数据实现最先进的预测性能
许多人工智能模型已被开发用于预测结直肠癌(CRC)的临床相关生物标志物,包括微卫星不稳定性(MSI)。然而,现有的深度学习网络需要大量的训练数据集,而这些数据集通常很难获得。在这项研究中,基于最新的使用移位窗口的分层视觉转换器(Swin Transformer [swan - t]),我们开发了一个有效的工作流程来预测CRC的生物标志物(MSI,高突变,染色体不稳定性,CpG岛甲基化表型,BRAF和TP53突变),需要相对较小的数据集。我们的swing -t工作流程在癌症基因组图谱结肠癌和直肠癌数据集(TCGA-CRC-DX)的研究内部交叉验证实验中基本实现了最先进的(SOTA)预测性能。在交叉研究外部验证中,它也表现出了出色的通用性,并且使用分子和细胞肿瘤学数据集进行训练(N = 1,065)和TCGA-CRC-DX (N = 462)进行测试,MSI在接受者工作特征曲线(AUROC)下的SOTA面积为0.90。最近的一项研究报告了类似的性能(AUROC = 0.91),在相同的测试数据集上使用了约8,000个训练样本(ResNet18)。swwin - t在使用小型训练数据集时非常高效,并且在200-500个训练样本中表现出稳健的预测性能。我们的研究结果表明,swwin - t可以比基于ResNet18和ShuffleNet的现有MSI预测算法效率高5-10倍。此外,swan - t模型证明了其准确预测MSI和BRAF突变状态的能力,从而可以在级联诊断工作流程中排除并减少后续标准测试之前的样本,从而减少周转时间和成本。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Pathology Clinical Research
Medicine-Pathology and Forensic Medicine
CiteScore
7.40
自引率
2.40%
发文量
47
审稿时长
20 weeks
期刊介绍:
The Journal of Pathology: Clinical Research and The Journal of Pathology serve as translational bridges between basic biomedical science and clinical medicine with particular emphasis on, but not restricted to, tissue based studies.
The focus of The Journal of Pathology: Clinical Research is the publication of studies that illuminate the clinical relevance of research in the broad area of the study of disease. Appropriately powered and validated studies with novel diagnostic, prognostic and predictive significance, and biomarker discover and validation, will be welcomed. Studies with a predominantly mechanistic basis will be more appropriate for the companion Journal of Pathology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


