KDR genetic predictor of toxicities induced by sorafenib and regorafenib
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 1
Abstract
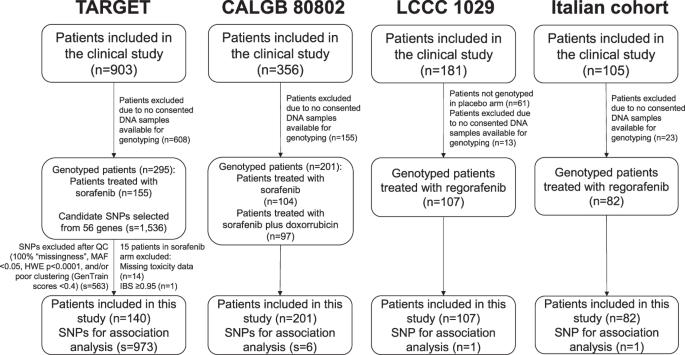
No biomarkers are available to predict toxicities induced by VEGFR TKIs. This study aimed to identify markers of toxicities induced by these drugs using a discovery-validation approach. The discovery set included 140 sorafenib-treated cancer patients (TARGET study) genotyped for SNPs in 56 genes. The most significant SNPs associated with grade ≥2 hypertension, diarrhea, dermatologic toxicities, and composite toxicity (any one of the toxicities) were tested for association with grade ≥2 toxicity in a validation set of 201 sorafenib-treated patients (Alliance/CALGB 80802). The validated SNP was tested for association with grade ≥2 toxicity in 107 (LCCC 1029) and 82 (Italian cohort) regorafenib-treated patients. SNP-toxicity associations were evaluated using logistic regression, and a meta-analysis between the studies was performed by inverse variance. Variant rs4864950 in KDR increased the risk of grade ≥2 composite toxicity in TARGET, Alliance/CALGB 80802, and the Italian cohort (meta-analysis p = 6.79 × 10−4, OR = 2.01, 95% CI 1.34–3.01). We identified a predictor of toxicities induced by VEGFR TKIs. NCT00073307 (TARGET), NCT01015833 (Alliance/CALGB 80802), and NCT01298570 (LCCC 1029).
索拉非尼和瑞戈非尼毒性的 KDR 遗传预测因子
目前还没有生物标志物可用于预测血管内皮生长因子受体 TKI 诱导的毒性。本研究采用发现-验证的方法,旨在确定这些药物诱发毒性的标志物。发现集包括140名索拉非尼治疗的癌症患者(TARGET研究),对56个基因中的SNP进行了基因分型。与≥2级高血压、腹泻、皮肤毒性和复合毒性(任何一种毒性)相关的最重要的SNPs在201名索拉非尼治疗患者(Alliance/CALGB 80802)组成的验证集中进行了与≥2级毒性相关性的测试。在107例(LCCC 1029)和82例(意大利队列)瑞戈非尼治疗患者中测试了已验证的SNP与≥2级毒性的相关性。利用逻辑回归评估了SNP与毒性的相关性,并通过反方差进行了研究间的荟萃分析。在TARGET、Alliance/CALGB 80802和意大利队列中,KDR中的变异体rs4864950增加了≥2级复合毒性的风险(荟萃分析p = 6.79 × 10-4,OR = 2.01,95% CI 1.34-3.01)。我们发现了一种 VEGFR TKIs 诱发毒性的预测因子。NCT00073307(TARGET)、NCT01015833(Alliance/CALGB 80802)和NCT01298570(LCCC 1029)。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


