Single-cell Sequencing Reveals Clearance of Blastula Chromosomal Mosaicism in In Vitro Fertilization Babies
Abstract
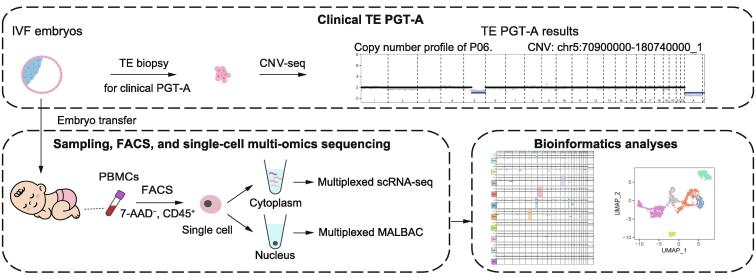
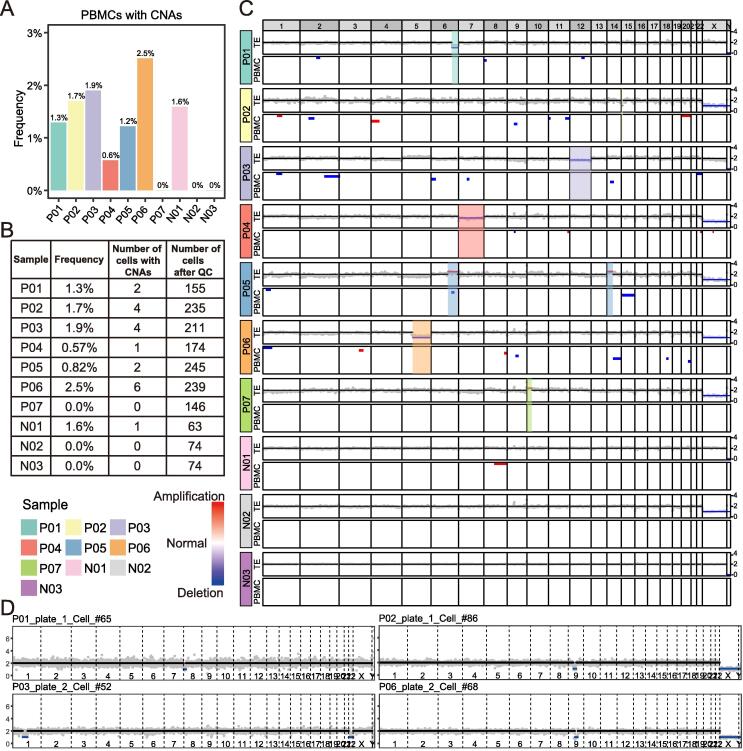
Although chromosomal mosaic embryos detected by trophectoderm (TE) biopsy offer healthy embryos available for transfer, high-resolution postnatal karyotyping and chromosome testing of the transferred embryos are insufficient. Here, we applied single-cell multi-omics sequencing for seven infants with blastula chromosomal mosaicism detected by TE biopsy. The chromosome ploidy was examined by single-cell genome analysis, with the cellular identity being identified by single-cell transcriptome analysis. A total of 1616 peripheral leukocytes from seven infants with embryonic chromosomal mosaicism and three control ones with euploid TE biopsy were analyzed. A small number of blood cells showed copy number alterations (CNAs) on seemingly random locations at a frequency of 0%−2.5% per infant. However, none of the cells showed CNAs that were the same as those of the corresponding TE biopsies. The blastula chromosomal mosaicism may be fully self-corrected, probably through the selective loss of the aneuploid cells during development, and the transferred embryos can be born as euploid infants without mosaic CNAs corresponding to the TE biopsies. The results provide a new reference for the evaluations of transferring chromosomal mosaic embryos in certain situations.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


