Insights into the interaction between the kusaginin and bovine serum albumin: Multi-spectroscopic techniques and computational approaches
IF 2.3
4区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 4
Abstract
Kusaginin, as a phenylethanoid glycoside, which has exhibited wide antioxidant and antimicrobial properties. The molecular mechanism underlying the broad biological activities of kusaginin has not yet been well documented. In this paper, the interaction of kusaginin with bovine serum albumin (BSA) has been explored by fluorescence spectra, UV‐vis absorption spectra, and circular dichroism (CD) spectra along with computational approaches. The fluorescence experiments showed that kusaginin could strongly quench the intrinsic fluorescence of BSA through both dynamic and static quenching mechanisms. The thermodynamic analysis suggested that hydrophobic force was the main force in stabilizing the BSA‐kusaginin complex. In addition, conformation changes of BSA were observed from three‐dimensional and synchronous fluorescence spectra, UV spectra, and CD spectra under experimental conditions. All these experimental results have been complemented and validated by the molecular docking and dynamic simulation studies, which revealed that kusaginin was bound on the hydrophobic cavity in subdomain IIA of BSA and formed a stable BSA‐kusaginin complex. Finally, density functional theory (DFT) calculation further implied that hydrogen bonds also support stabilizing the BSA‐kusaginin complex. This research may aid in understanding the pharmacological characteristics of kusaginin and provide a vital reference modeling for the design of analogues drugs.
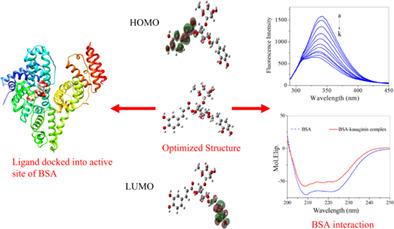
洞察草蓟素和牛血清白蛋白之间的相互作用:多光谱技术和计算方法
草蓟素作为一种苯基乙醇苷类化合物,具有广泛的抗氧化和抗菌性能。草蓟素广泛生物活性的分子机制尚未得到很好的证实。本文采用荧光光谱、紫外-可见吸收光谱和圆二色性(CD)光谱并结合计算方法研究了草蓟素与牛血清白蛋白(BSA)的相互作用。荧光实验表明,草蓟素可以通过动态和静态猝灭机制强烈猝灭牛血清白蛋白的固有荧光。热力学分析表明疏水力是稳定bsa -草草素配合物的主要力量。此外,在实验条件下,通过三维和同步荧光光谱、紫外光谱和CD光谱观察了牛血清白蛋白的构象变化。这些实验结果都得到了分子对接和动态模拟研究的补充和验证,发现草蓟素结合在牛血清白蛋白IIA亚结构域的疏水空腔上,形成了稳定的牛血清白蛋白-草蓟素复合物。最后,密度泛函理论(DFT)计算进一步表明,氢键也支持稳定BSA-kusaginin配合物。本研究有助于了解草蓟素的药理特性,并为其类似物的设计提供重要的参考模型。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Recognition
生物-生化与分子生物学
CiteScore
4.60
自引率
3.70%
发文量
68
审稿时长
2.7 months
期刊介绍:
Journal of Molecular Recognition (JMR) publishes original research papers and reviews describing substantial advances in our understanding of molecular recognition phenomena in life sciences, covering all aspects from biochemistry, molecular biology, medicine, and biophysics. The research may employ experimental, theoretical and/or computational approaches.
The focus of the journal is on recognition phenomena involving biomolecules and their biological / biochemical partners rather than on the recognition of metal ions or inorganic compounds. Molecular recognition involves non-covalent specific interactions between two or more biological molecules, molecular aggregates, cellular modules or organelles, as exemplified by receptor-ligand, antigen-antibody, nucleic acid-protein, sugar-lectin, to mention just a few of the possible interactions. The journal invites manuscripts that aim to achieve a complete description of molecular recognition mechanisms between well-characterized biomolecules in terms of structure, dynamics and biological activity. Such studies may help the future development of new drugs and vaccines, although the experimental testing of new drugs and vaccines falls outside the scope of the journal. Manuscripts that describe the application of standard approaches and techniques to design or model new molecular entities or to describe interactions between biomolecules, but do not provide new insights into molecular recognition processes will not be considered. Similarly, manuscripts involving biomolecules uncharacterized at the sequence level (e.g. calf thymus DNA) will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


