Adding context to the pneumococcal core genes using bioinformatic analysis of the intergenic pangenome of Streptococcus pneumoniae.
IF 2.8
Q2 MATHEMATICAL & COMPUTATIONAL BIOLOGY
引用次数: 0
Abstract
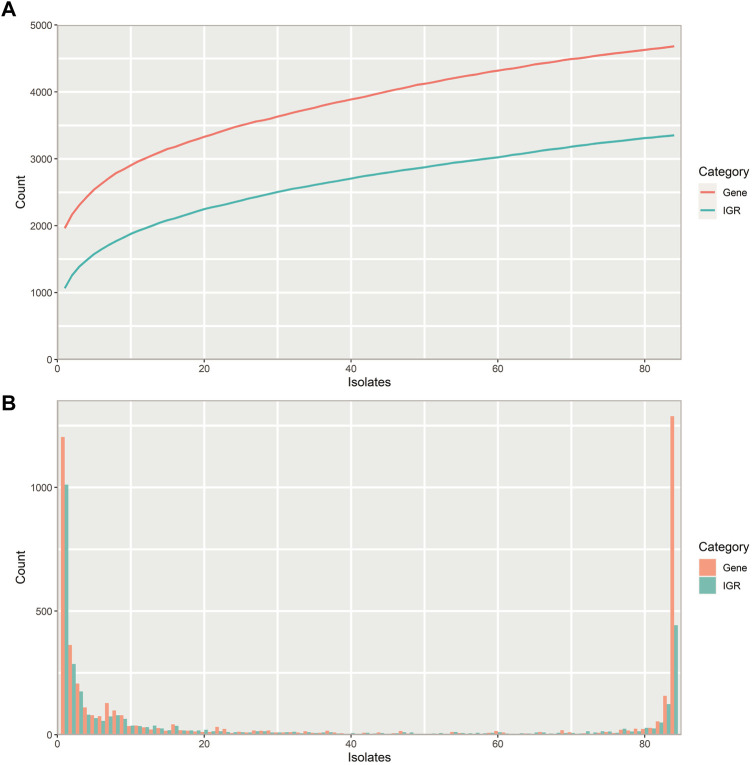
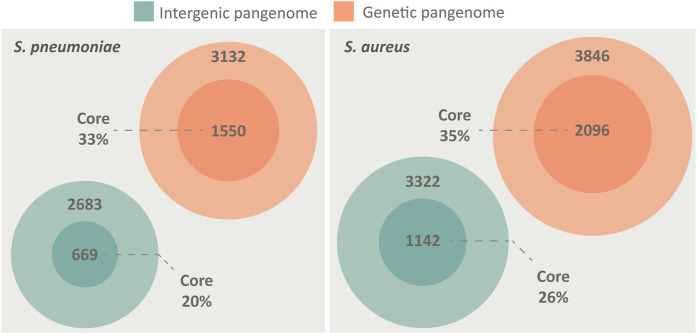
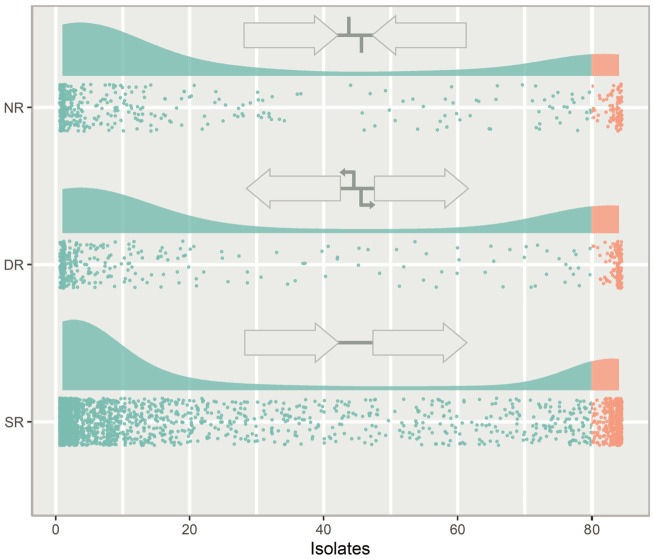
Whole genome sequencing offers great opportunities for linking genotypes to phenotypes aiding in our understanding of human disease and bacterial pathogenicity. However, these analyses often overlook non-coding intergenic regions (IGRs). By disregarding the IGRs, crucial information is lost, as genes have little biological function without expression. In this study, we present the first complete pangenome of the important human pathogen Streptococcus pneumoniae (pneumococcus), spanning both the genes and IGRs. We show that the pneumococcus species retains a small core genome of IGRs that are present across all isolates. Gene expression is highly dependent on these core IGRs, and often several copies of these core IGRs are found across each genome. Core genes and core IGRs show a clear linkage as 81% of core genes are associated with core IGRs. Additionally, we identify a single IGR within the core genome that is always occupied by one of two highly distinct sequences, scattered across the phylogenetic tree. Their distribution indicates that this IGR is transferred between isolates through horizontal regulatory transfer independent of the flanking genes and that each type likely serves different regulatory roles depending on their genetic context.
利用肺炎链球菌基因间泛基因组的生物信息学分析为肺炎球菌核心基因添加背景。
引言:全基因组测序为将基因型与表型联系起来提供了巨大的机会,有助于我们了解人类疾病和细菌致病性。然而,这些分析往往忽略了非编码基因间区(IGRs)。如果忽略igr,关键信息就会丢失,因为没有表达的基因几乎没有生物学功能。方法/结果:在这项研究中,我们首次获得了人类重要病原体肺炎链球菌(肺炎球菌)的完整泛基因组,涵盖了基因和IGRs。我们表明,肺炎球菌物种保留了igr的小核心基因组,存在于所有分离株中。基因表达高度依赖于这些核心igr,并且通常在每个基因组中发现这些核心igr的几个拷贝。核心基因和核心IGRs有明显的联系,81%的核心基因与核心IGRs相关。此外,我们在核心基因组中发现了一个单一的IGR,该IGR总是由两个高度不同的序列之一占据,分散在整个系统发育树中。讨论:它们的分布表明,这种IGR通过独立于侧翼基因的水平调控转移在分离株之间转移,并且每种类型可能根据其遗传背景发挥不同的调控作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


