Association of ITPA gene polymorphisms with adverse effects of AZA/6-MP administration: a systematic review and meta-analysis
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 7
Abstract
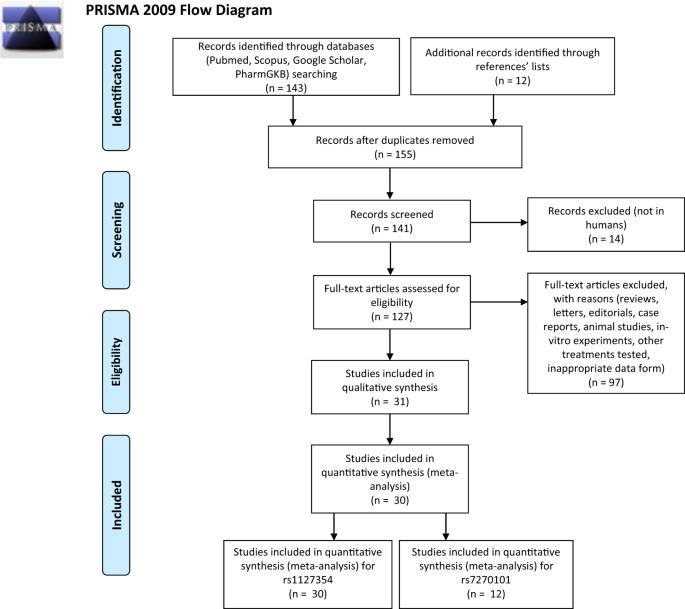
Azathioprine (AZA) and its metabolite, mercaptopurine (6-MP), are widely used immunosuppressant drugs. Polymorphisms in genes implicated in AZA/6-MP metabolism, reportedly, could account in part for their potential toxicity. In the present study we performed a systematic review and a meta-analysis, comprising 30 studies and 3582 individuals, to investigate the putative genetic association of two inosine triphosphatase (ITPA) polymorphisms with adverse effects in patients treated with AZA/6-MP. We found that rs1127354 is associated with neutropenia in general populations and in children (OR: 2.39, 95%CI: 1.97–2.90, and OR: 2.43, 95%CI: 2.12–2.79, respectively), and with all adverse effects tested herein in adult populations (OR: 2.12, 95%CI: 1.22–3.69). We also found that rs7270101 is associated with neutropenia and leucopenia in all-ages populations (OR: 2.93, 95%CI: 2.36–3.63, and OR: 2.82, 95%CI: 1.76–4.50, respectively) and with all adverse effects tested herein in children (OR: 1.74, 95%CI: 1.06–2.87). Stratification according to background disease, in combination with multiple comparisons corrections, verified neutropenia to be associated with both polymorphisms, in acute lymphoblastic leukemia (ALL) patients. These findings suggest that ITPA polymorphisms could be used as predictive biomarkers for adverse effects of thiopurine drugs to eliminate intolerance in ALL patients and clarify dosing in patients with different ITPA variants.
ITPA 基因多态性与 AZA/6-MP 给药不良反应的关系:系统回顾和荟萃分析
硫唑嘌呤(AZA)及其代谢产物巯基嘌呤(6-MP)是广泛使用的免疫抑制剂。据报道,与 AZA/6-MP 代谢有关的基因多态性可能是造成其潜在毒性的部分原因。在本研究中,我们对 30 项研究和 3582 人进行了系统回顾和荟萃分析,以调查两种肌苷三磷酸酶(ITPA)多态性与接受 AZA/6-MP 治疗的患者不良反应之间的假定遗传关联。我们发现,在普通人群和儿童中,rs1127354 与中性粒细胞减少症相关(OR:2.39,95%CI:1.97-2.90;OR:2.43,95%CI:2.12-2.79),在成人人群中,rs1127354 与本文测试的所有不良反应相关(OR:2.12,95%CI:1.22-3.69)。我们还发现,在所有年龄段人群中,rs7270101 与中性粒细胞减少症和白细胞减少症相关(OR:2.93,95%CI:2.36-3.63;OR:2.82,95%CI:1.76-4.50),在儿童中则与本文测试的所有不良反应相关(OR:1.74,95%CI:1.06-2.87)。在急性淋巴细胞白血病(ALL)患者中,根据背景疾病进行分层并结合多重比较校正,证实了中性粒细胞减少与这两种多态性有关。这些研究结果表明,ITPA多态性可作为硫嘌呤类药物不良反应的预测性生物标记物,以消除ALL患者的不耐受性,并明确不同ITPA变体患者的用药剂量。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


