Subcellular Proteomic Analysis Reveals Dysregulation in Organization of Human A549 Cells Infected with Influenza Virus H7N9
IF 0.5
4区 生物学
Q4 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
H7N9 influenza virus poses a high risk to human beings and proteomic evaluations of these infections may help to better understand its pathogenic mechanisms in human systems. Objective: To find membrane proteins related to H7N9 infection. Here, we infected primary human alveolar adenocarcinoma epithelial cells (A549) cells with H7N9 (including wild and mutant strains) and then produced enriched cellular membrane isolations which were evaluated by western blot. The proteins in these cell membrane fractions were analyzed using the isobaric Tags for Relative and Absolute Quantitation (iTRAQ) proteome technologies. Differentially expressed proteins (n = 32) were identified following liquid chromatography-tandem mass spectrometry, including 20 down-regulated proteins such as CD44 antigen, and CD151 antigen, and 12 up-regulated proteins such as tight junction protein ZO-1, and prostaglandin reductase 1. Gene Ontology database searching revealed that 20 out of the 32 differentially expressed proteins were localized to the plasma membrane. These proteins were primarily associated with cellular component organization (n = 20), and enriched in the Reactome pathway of extracellular matrix organization (n = 4). These findings indicate that H7N9 may dysregulate cellular organization via specific alterations to the protein profile of the plasma membrane.
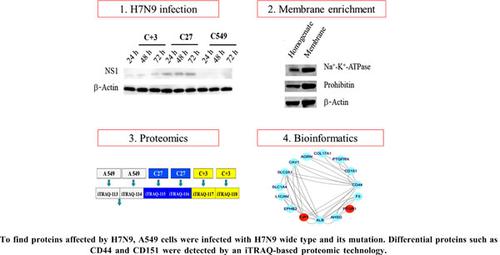
亚细胞蛋白质组学分析揭示感染H7N9流感病毒的人A549细胞组织失调
H7N9流感病毒对人类构成高风险,对这些感染进行蛋白质组学评估可能有助于更好地了解其在人体系统中的致病机制。目的:寻找与H7N9感染相关的膜蛋白。我们用H7N9(包括野生株和突变株)感染原代人肺泡腺癌上皮细胞(A549),然后获得富集的细胞膜分离物,并用western blot对其进行评价。利用等压标记相对和绝对定量(iTRAQ)蛋白质组技术对这些细胞膜组分中的蛋白质进行分析。采用液相色谱-串联质谱法鉴定差异表达蛋白(n = 32),其中CD44抗原、CD151抗原等下调蛋白20个,紧密连接蛋白ZO-1、前列腺素还原酶1等上调蛋白12个。基因本体数据库检索显示,32个差异表达蛋白中有20个定位在质膜上。这些蛋白主要与细胞组分组织相关(n = 20),并富集于细胞外基质组织的Reactome通路(n = 4)。这些发现表明H7N9可能通过特异性改变质膜蛋白谱来调节细胞组织。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current Proteomics
BIOCHEMICAL RESEARCH METHODS-BIOCHEMISTRY & MOLECULAR BIOLOGY
CiteScore
1.60
自引率
0.00%
发文量
25
审稿时长
>0 weeks
期刊介绍:
Research in the emerging field of proteomics is growing at an extremely rapid rate. The principal aim of Current Proteomics is to publish well-timed in-depth/mini review articles in this fast-expanding area on topics relevant and significant to the development of proteomics. Current Proteomics is an essential journal for everyone involved in proteomics and related fields in both academia and industry.
Current Proteomics publishes in-depth/mini review articles in all aspects of the fast-expanding field of proteomics. All areas of proteomics are covered together with the methodology, software, databases, technological advances and applications of proteomics, including functional proteomics. Diverse technologies covered include but are not limited to:
Protein separation and characterization techniques
2-D gel electrophoresis and image analysis
Techniques for protein expression profiling including mass spectrometry-based methods and algorithms for correlative database searching
Determination of co-translational and post- translational modification of proteins
Protein/peptide microarrays
Biomolecular interaction analysis
Analysis of protein complexes
Yeast two-hybrid projects
Protein-protein interaction (protein interactome) pathways and cell signaling networks
Systems biology
Proteome informatics (bioinformatics)
Knowledge integration and management tools
High-throughput protein structural studies (using mass spectrometry, nuclear magnetic resonance and X-ray crystallography)
High-throughput computational methods for protein 3-D structure as well as function determination
Robotics, nanotechnology, and microfluidics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


