Landscape genomics reveals adaptive genetic differentiation driven by multiple environmental variables in naked barley on the Qinghai-Tibetan Plateau
IF 3.1
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
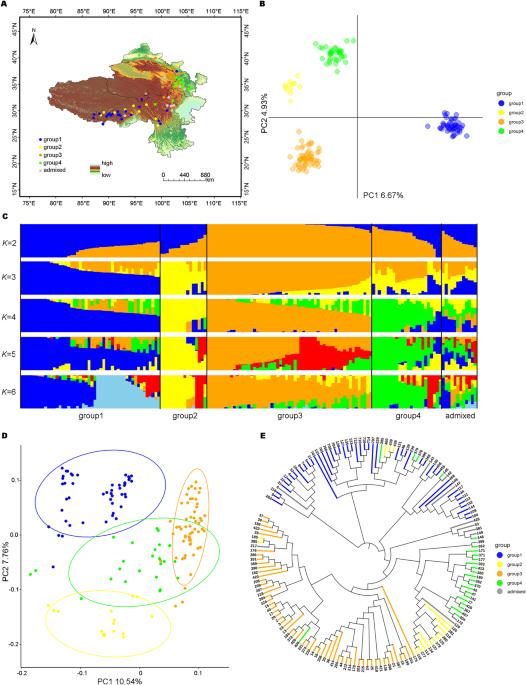
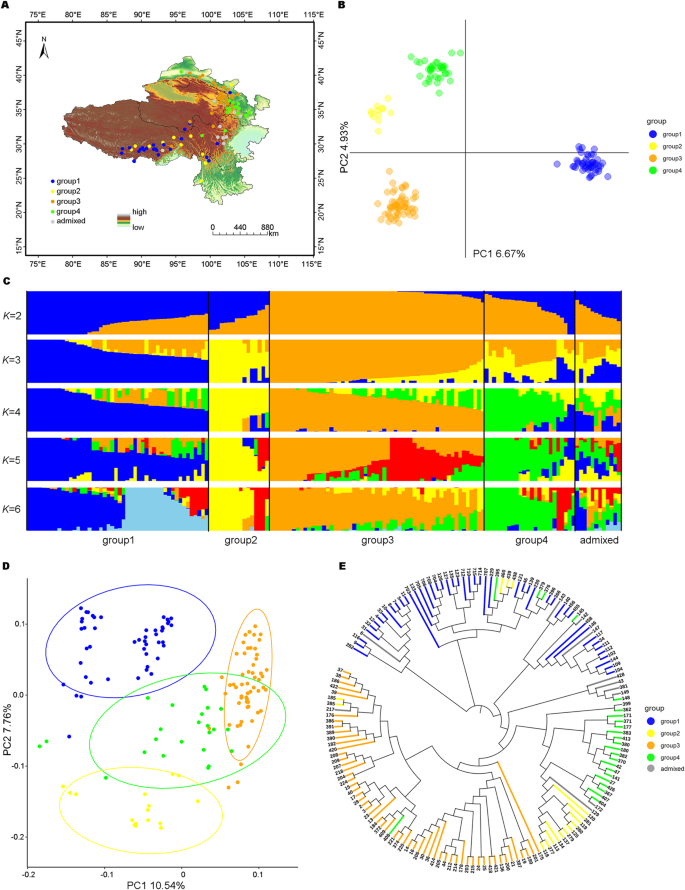
Understanding the local adaptation of crops has long been a concern of evolutionary biologists and molecular ecologists. Identifying the adaptive genetic variability in the genome is crucial not only to provide insights into the genetic mechanism of local adaptation but also to explore the adaptation potential of crops. This study aimed to identify the climatic drivers of naked barley landraces and putative adaptive loci driving local adaptation on the Qinghai-Tibetan Plateau (QTP). To this end, a total of 157 diverse naked barley accessions were genotyped using the genotyping-by-sequencing approach, which yielded 3123 high-quality SNPs for population structure analysis and partial redundancy analysis, and 37,636 SNPs for outlier analysis. The population structure analysis indicated that naked barley landraces could be divided into four groups. We found that the genomic diversity of naked barley landraces could be partly traced back to the geographical and environmental diversity of the landscape. In total, 136 signatures associated with temperature, precipitation, and ultraviolet radiation were identified, of which 13 had pleiotropic effects. We mapped 447 genes, including a known gene HvSs1. Some genes involved in cold stress and regulation of flowering time were detected near eight signatures. Taken together, these results highlight the existence of putative adaptive loci in naked barley on QTP and thus improve our current understanding of the genetic basis of local adaptation.
景观基因组学揭示了青藏高原裸大麦在多种环境变量驱动下的适应性遗传分化。
长期以来,了解作物的局部适应一直是进化生物学家和分子生态学家关注的问题。识别基因组中的适应性遗传变异不仅对深入了解局部适应的遗传机制至关重要,而且对探索作物的适应潜力也至关重要。本研究旨在确定青藏高原裸大麦地方品种的气候驱动因素和驱动当地适应的假定适应性基因座(QTP)。为此,使用测序基因分型方法对157份不同的裸大麦材料进行了基因分型,共产生3123个高质量SNPs用于群体结构分析和部分冗余分析,37636个SNPs用于异常值分析。群体结构分析表明,裸大麦地方品种可分为四个类群。我们发现裸大麦地方品种的基因组多样性可以部分追溯到景观的地理和环境多样性。总共鉴定出136个与温度、降水和紫外线辐射有关的特征,其中13个具有多效性效应。我们定位了447个基因,包括一个已知的基因HvSs1。在八个特征附近检测到一些与冷胁迫和开花时间调控有关的基因。总之,这些结果突出了在QTP上裸大麦中假定的适应性基因座的存在,从而提高了我们目前对局部适应性遗传基础的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


