IBD sharing patterns as intra-breed admixture indicators in small ruminants
IF 3.1
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
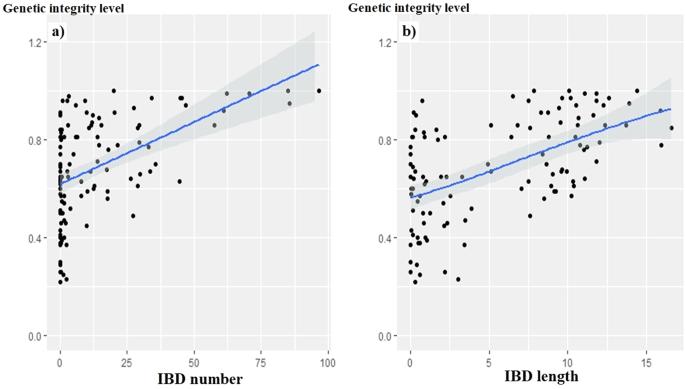
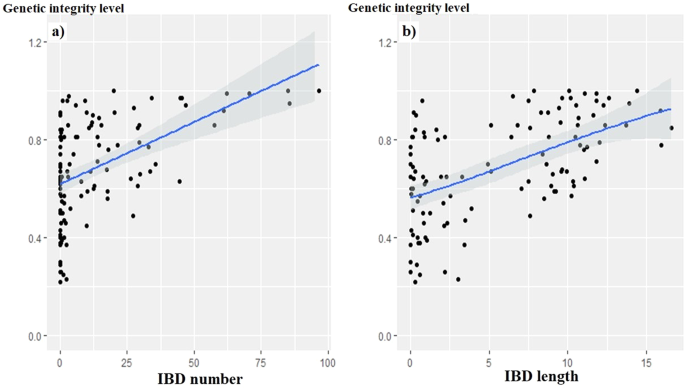
In this study, we investigated how IBD patterns shared between individuals of the same breed could be informative of its admixture level, with the underlying assumption that the most admixed breeds, i.e. the least genetically isolated, should have a much more fragmented genome. We considered 111 goat breeds (i.e. 2501 individuals) and 156 sheep breeds (i.e. 3304 individuals) from Europe, Africa and Asia, for which beadchip SNP genotypes had been performed. We inferred the breed’s level of admixture from: (i) the proportion of the genome shared by breed’s members (i.e. “genetic integrity level” assessed from ADMIXTURE software analyses), and (ii) the “AV index” (calculated from Reynolds’ genetic distances), used as a proxy for the “genetic distinctiveness”. In both goat and sheep datasets, the statistical analyses (comparison of means, Spearman correlations, LM and GAM models) revealed that the most genetically isolated breeds, also showed IBD profiles made up of more shared IBD segments, which were also longer. These results pave the way for further research that could lead to the development of admixture indicators, based on the characterization of intra-breed shared IBD segments, particularly effective as they would be independent of the knowledge of the whole genetic landscape in which the breeds evolve. Finally, by highlighting the fragmentation experienced by the genomes subjected to crossbreeding carried out over the last few generations, the study reminds us of the need to preserve local breeds and the integrity of their adaptive architectures that have been shaped over the centuries.
小反刍动物IBD共享模式作为种内混合指标。
在这项研究中,我们调查了同一品种的个体之间共享的IBD模式如何能够为其混合水平提供信息,其基本假设是,最混合的品种,即遗传隔离最少的品种,应该有一个更碎片化的基因组。我们考虑了来自欧洲、非洲和亚洲的111个山羊品种(即2501个个体)和156个绵羊品种(即3304个个体),对其进行了珠芯片SNP基因型分析。我们从以下方面推断出该品种的混合水平:(i)品种成员共享的基因组比例(即admixture软件分析评估的“遗传完整性水平”),以及(ii)“AV指数”(根据Reynolds的遗传距离计算),用作“遗传特异性”的代表。在山羊和绵羊的数据集中,统计分析(平均值比较、Spearman相关性、LM和GAM模型)显示,遗传上最孤立的品种也显示出由更多共享的IBD片段组成的IBD图谱,这些片段也更长。这些结果为进一步的研究铺平了道路,这些研究可能会导致基于种内共享IBD片段特征的混合指标的开发,特别有效,因为它们独立于品种进化的整个遗传景观的知识。最后,通过强调过去几代进行杂交的基因组所经历的碎片化,这项研究提醒我们有必要保护当地品种及其几个世纪以来形成的适应性结构的完整性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


