Comparison of serum fractionation methods by data independent label-free proteomics
Q4 Biochemistry, Genetics and Molecular Biology
引用次数: 5
Abstract
Off-line sample prefractionations applied prior to biomarker discovery proteomics are options to enable more protein identifications and detect low-abundance proteins. This work compared five commercial methods efficiency to raw serum analysis using label-free proteomics. The variability of the protein quantities determined for each process was similar to the unprefractionated serum. A 49% increase in protein identifications and 12.2% of reliable quantification were obtained. A 61 times lower limit of protein quantitation was reached compared to protein concentrations observed in raw serum. The concentrations of detected proteins were confronted to estimated reference values.
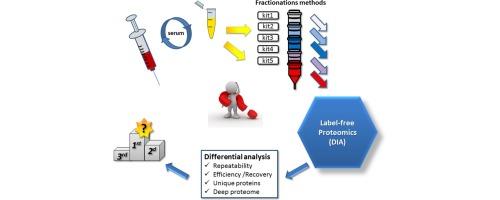
数据独立的无标记蛋白质组学血清分离方法的比较
在生物标志物发现之前进行离线样品预分离,蛋白质组学是实现更多蛋白质鉴定和检测低丰度蛋白质的选择。这项工作比较了五种商业方法与使用无标记蛋白质组学的原始血清分析的效率。每种工艺测定的蛋白质量的可变性与未预分离的血清相似。获得了49%的蛋白质鉴定和12.2%的可靠定量。与原血清中观察到的蛋白质浓度相比,达到了蛋白质定量下限的61倍。检测到的蛋白质浓度与估计的参考值相符。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

EuPA Open Proteomics
Biochemistry, Genetics and Molecular Biology-Biochemistry
自引率
0.00%
发文量
0
审稿时长
103 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


