Genomic and transcriptomic insights into complex virus–prokaryote interactions in marine biofilms
IF 10.8
1区 环境科学与生态学
Q1 ECOLOGY
引用次数: 0
Abstract
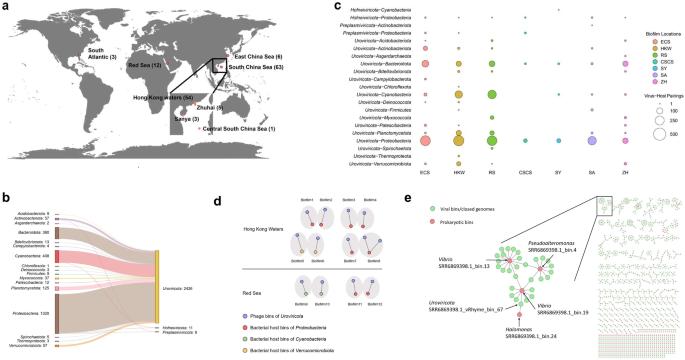
Marine biofilms are complex communities of microorganisms that play a crucial ecological role in oceans. Although prokaryotes are the dominant members of these biofilms, little is known about their interactions with viruses. By analysing publicly available and newly sequenced metagenomic data, we identified 2446 virus–prokaryote connections in 84 marine biofilms. Most of these connections were between the bacteriophages in the Uroviricota phylum and the bacteria of Proteobacteria, Cyanobacteria and Bacteroidota. The network of virus–host pairs is complex; a single virus can infect multiple prokaryotic populations or a single prokaryote is susceptible to several viral populations. Analysis of genomes of paired prokaryotes and viruses revealed the presence of 425 putative auxiliary metabolic genes (AMGs), 239 viral genes related to restriction–modification (RM) systems and 38,538 prokaryotic anti-viral defence-related genes involved in 15 defence systems. Transcriptomic evidence from newly established biofilms revealed the expression of viral genes, including AMGs and RM, and prokaryotic defence systems, indicating the active interplay between viruses and prokaryotes. A comparison between biofilms and seawater showed that biofilm prokaryotes have more abundant defence genes than seawater prokaryotes, and the defence gene composition differs between biofilms and the surrounding seawater. Overall, our study unveiled active viruses in natural biofilms and their complex interplay with prokaryotes, which may result in the blooming of defence strategists in biofilms. The detachment of bloomed defence strategists may reduce the infectivity of viruses in seawater and result in the emergence of a novel role of marine biofilms.
海洋生物膜中复杂病毒-原核生物相互作用的基因组学和转录组学研究。
海洋生物膜是复杂的微生物群落,在海洋中发挥着至关重要的生态作用。尽管原核生物是这些生物膜的主要成员,但人们对它们与病毒的相互作用知之甚少。通过分析公开的和新测序的宏基因组数据,我们在84个海洋生物膜中鉴定了2446个病毒原核生物连接。这些联系大多发生在绿病毒门的噬菌体与变形菌门、蓝细菌门和拟杆菌门的细菌之间。病毒-宿主对的网络是复杂的;单个病毒可以感染多个原核生物群体,或者单个原核生物对多个病毒群体敏感。对配对原核生物和病毒的基因组分析显示,存在425个推定的辅助代谢基因(AMG)、239个与限制性修饰(RM)系统相关的病毒基因和38538个参与15个防御系统的原核抗病毒防御相关基因。来自新建立的生物膜的转录组学证据揭示了病毒基因的表达,包括AMG和RM,以及原核生物防御系统,表明病毒和原核生物之间的积极相互作用。生物膜和海水的比较表明,生物膜原核生物比海水原核生物具有更丰富的防御基因,并且生物膜和周围海水的防御基因组成不同。总的来说,我们的研究揭示了天然生物膜中的活性病毒及其与原核生物的复杂相互作用,这可能导致生物膜中防御战略家的蓬勃发展。成熟的防御战略家的分离可能会降低海水中病毒的传染性,并导致海洋生物膜的新作用的出现。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ISME Journal
环境科学-生态学
CiteScore
22.10
自引率
2.70%
发文量
171
审稿时长
2.6 months
期刊介绍:
The ISME Journal covers the diverse and integrated areas of microbial ecology. We encourage contributions that represent major advances for the study of microbial ecosystems, communities, and interactions of microorganisms in the environment. Articles in The ISME Journal describe pioneering discoveries of wide appeal that enhance our understanding of functional and mechanistic relationships among microorganisms, their communities, and their habitats.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


