Construction of a 3D whole organism spatial atlas by joint modelling of multiple slices with deep neural networks
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Spatial transcriptomics (ST) technologies are revolutionizing the way to explore the spatial architecture of tissues. Currently, ST data analysis is often restricted to a single two-dimensional (2D) tissue slice, limiting our capacity to understand biological processes that take place in 3D space. Here we present STitch3D, a unified framework that integrates multiple ST slices to reconstruct 3D cellular structures. By jointly modelling multiple slices and integrating them with single-cell RNA-sequencing data, STitch3D simultaneously identifies 3D spatial regions with coherent gene-expression levels and reveals 3D cell-type distributions. STitch3D distinguishes biological variation among slices from batch effects, and effectively borrows information across slices to assemble powerful 3D models. Through comprehensive experiments, we demonstrate STitch3D’s performance in building comprehensive 3D architectures, which allow 3D analysis in the entire tissue region or even the whole organism. The outputs of STitch3D can be used for multiple downstream tasks, enabling a comprehensive understanding of biological systems. Computational methods for analysing single 2D tissue slices from spatial transcriptomics studies are well established, but their extension to the 3D domain is challenging. Wang et al. develop a deep learning framework that can perform 3D reconstruction of cellular structures in tissues as well as whole organisms.
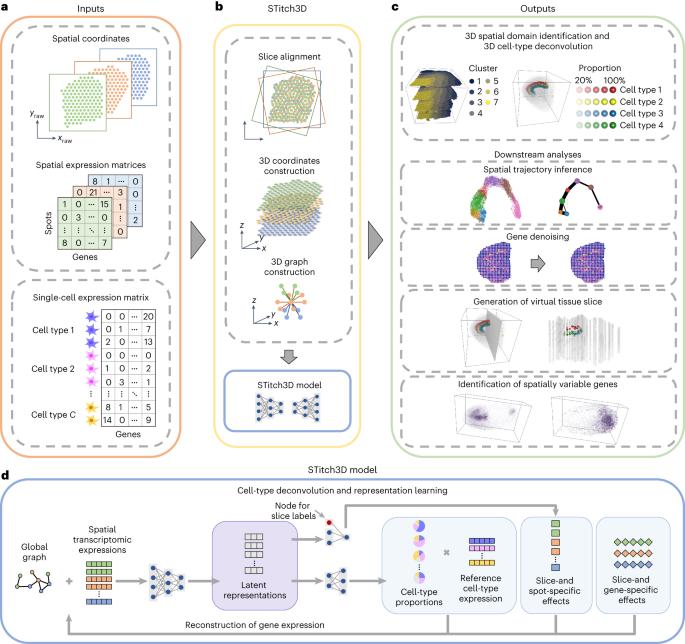
利用深度神经网络对多个切片进行联合建模,构建三维全生物体空间图谱
空间转录组学(ST)技术正在彻底改变探索组织空间结构的方式。目前,空间转录组学数据分析往往局限于单一的二维(2D)组织切片,这限制了我们了解三维空间生物过程的能力。在这里,我们提出了 STitch3D,这是一个整合多个 ST 切片以重建三维细胞结构的统一框架。通过对多个切片进行联合建模并将其与单细胞 RNA 序列数据整合,STitch3D 可同时识别具有一致基因表达水平的三维空间区域,并揭示三维细胞类型分布。STitch3D 能将切片间的生物变异与批次效应区分开来,并有效地借用切片间的信息来建立功能强大的三维模型。通过综合实验,我们证明了 STitch3D 在构建综合三维架构方面的性能,从而可以对整个组织区域甚至整个生物体进行三维分析。STitch3D 的输出结果可用于多种下游任务,从而实现对生物系统的全面了解。用于分析空间转录组学研究中的单个二维组织切片的计算方法已经成熟,但将其扩展到三维领域却具有挑战性。Wang 等人开发了一种深度学习框架,可以对组织和整个生物体的细胞结构进行三维重建。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


