Mitigating the missing-fragmentation problem in de novo peptide sequencing with a two-stage graph-based deep learning model
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
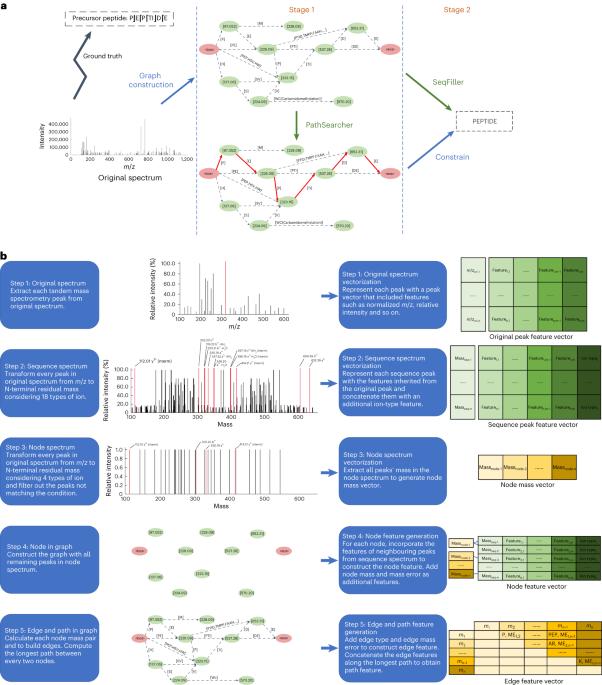
Novel protein discovery and immunopeptidomics depend on highly sensitive de novo peptide sequencing with tandem mass spectrometry. Despite notable improvement using deep learning models, the missing-fragmentation problem remains an important hurdle that severely degrades the performance of de novo peptide sequencing. Here we reveal that in the process of peptide prediction, missing fragmentation results in the generation of incorrect amino acids within those regions and causes error accumulation thereafter. To tackle this problem, we propose GraphNovo, a two-stage de novo peptide-sequencing algorithm based on a graph neural network. GraphNovo focuses on finding the optimal path in the first stage to guide the sequence prediction in the second stage. Our experiments demonstrate that GraphNovo mitigates the effects of missing fragmentation and outperforms the state-of-the-art de novo peptide-sequencing algorithms. Identifying unknown peptides in tandem mass spectrometry is challenging as fragmentation of precursor peptides can be incomplete. Mao and colleagues present a method based on graph neural networks and a path-searching model to create more stable sequence predictions.
利用基于图的两阶段深度学习模型缓解全新多肽测序中的缺失-碎片问题
新蛋白质的发现和免疫肽组学依赖于串联质谱法的高灵敏度从头肽测序。尽管利用深度学习模型取得了显著的改进,但缺失片段问题仍然是严重降低从头肽测序性能的一个重要障碍。在此,我们揭示了在肽段预测过程中,缺失片段会导致在这些区域生成错误的氨基酸,并造成此后的错误累积。为解决这一问题,我们提出了基于图神经网络的两阶段肽测序算法 GraphNovo。GraphNovo 专注于在第一阶段找到最优路径,以指导第二阶段的序列预测。我们的实验证明,GraphNovo 可减轻缺失片段的影响,其性能优于最先进的肽测序算法。在串联质谱中识别未知肽具有挑战性,因为前体肽的碎片可能不完整。Mao 及其同事提出了一种基于图神经网络和路径搜索模型的方法,以创建更稳定的序列预测。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


