Rapid Identification of Protein-Protein Interactions in Plants
Q1 Agricultural and Biological Sciences
Youjun Zhang, Roberto Natale, Adilson Pereira Domingues Júnior, Mitchell Rey Toleco, Beata Siemiatkowska, Norma Fàbregas, Alisdair R. Fernie
下载PDF
{"title":"Rapid Identification of Protein-Protein Interactions in Plants","authors":"Youjun Zhang, Roberto Natale, Adilson Pereira Domingues Júnior, Mitchell Rey Toleco, Beata Siemiatkowska, Norma Fàbregas, Alisdair R. Fernie","doi":"10.1002/cppb.20099","DOIUrl":null,"url":null,"abstract":"<p>Enzyme-enzyme interactions can be discovered by affinity purification mass spectrometry (AP-MS) under in vivo conditions. Tagged enzymes can either be transiently transformed into plant leaves or stably transformed into plant cells prior to AP-MS. The success of AP-MS depends on the levels and stability of the bait protein, the stability of the protein-protein interactions, and the efficiency of trypsin digestion and recovery of tryptic peptides for MS analysis. Unlike in-gel-digestion AP-MS, in which the gel is cut into pieces for several independent trypsin digestions, we uses a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification. Thus, a single replicate within an AP-MS experiment constitutes a single sample for LC-MS measurement. In subsequent data analysis, normalized signal intensities can be processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software. Following analysis of co-sublocalization of “bait” and “prey,” we suggest considering only the protein pairs for which the intensities were more than 2% compared with the bait, corresponding to FC-A values of at least four within-biological replicates, which we recommend as minimum. If the procedure is faithfully followed, experimental assessment of enzyme-enzyme interactions can be carried out in Arabidopsis within 3 weeks (transient expression) or 5 weeks (stable expression). © 2019 The Authors.</p><p><b>Basic Protocol 1</b>: Gene cloning to the destination vectors</p><p><b>Alternate Protocol</b>: In-Fusion or Gibson gene cloning protocol</p><p><b>Basic Protocol 2</b>: Transformation of baits into the plant cell culture or plant leaf</p><p><b>Basic Protocol 3</b>: Affinity purification of protein complexes</p><p><b>Basic Protocol 4</b>: On-bead trypsin/LysC digestion and C18 column peptide desalting and concentration</p><p><b>Basic Protocol 5</b>: Data analysis and quality control</p>","PeriodicalId":10932,"journal":{"name":"Current protocols in plant biology","volume":"4 4","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2019-11-12","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cppb.20099","citationCount":"19","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in plant biology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cppb.20099","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Agricultural and Biological Sciences","Score":null,"Total":0}
引用次数: 19
引用
批量引用
Abstract
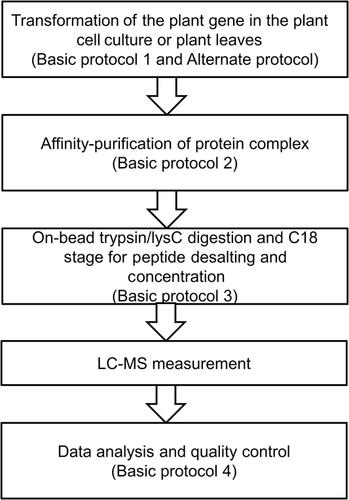
Enzyme-enzyme interactions can be discovered by affinity purification mass spectrometry (AP-MS) under in vivo conditions. Tagged enzymes can either be transiently transformed into plant leaves or stably transformed into plant cells prior to AP-MS. The success of AP-MS depends on the levels and stability of the bait protein, the stability of the protein-protein interactions, and the efficiency of trypsin digestion and recovery of tryptic peptides for MS analysis. Unlike in-gel-digestion AP-MS, in which the gel is cut into pieces for several independent trypsin digestions, we uses a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification. Thus, a single replicate within an AP-MS experiment constitutes a single sample for LC-MS measurement. In subsequent data analysis, normalized signal intensities can be processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software. Following analysis of co-sublocalization of “bait” and “prey,” we suggest considering only the protein pairs for which the intensities were more than 2% compared with the bait, corresponding to FC-A values of at least four within-biological replicates, which we recommend as minimum. If the procedure is faithfully followed, experimental assessment of enzyme-enzyme interactions can be carried out in Arabidopsis within 3 weeks (transient expression) or 5 weeks (stable expression). © 2019 The Authors.
Basic Protocol 1 : Gene cloning to the destination vectors
Alternate Protocol : In-Fusion or Gibson gene cloning protocol
Basic Protocol 2 : Transformation of baits into the plant cell culture or plant leaf
Basic Protocol 3 : Affinity purification of protein complexes
Basic Protocol 4 : On-bead trypsin/LysC digestion and C18 column peptide desalting and concentration
Basic Protocol 5 : Data analysis and quality control
植物中蛋白质-蛋白质相互作用的快速鉴定
酶-酶的相互作用可以通过亲和纯化质谱(AP-MS)在体内条件下发现。在AP-MS之前,标记的酶可以瞬间转化为植物叶片,也可以稳定地转化为植物细胞。AP-MS的成功取决于诱饵蛋白的水平和稳定性,蛋白-蛋白相互作用的稳定性,以及胰蛋白酶消化和用于MS分析的胰蛋白酶肽的回收效率。与凝胶消解AP-MS不同,凝胶被切成几块用于几个独立的胰蛋白酶消解,我们使用基于蛋白质组学的溶液消解方法,在亲和纯化后直接消解珠子上的蛋白质。因此,AP-MS实验中的单个重复构成LC-MS测量的单个样品。在随后的数据分析中,可以使用CRAPome软件中嵌入的SAINT算法对归一化信号强度进行处理,以确定fold-change abundance (FC-A)评分。在对“诱饵”和“猎物”的共亚定位进行分析后,我们建议只考虑与诱饵相比强度大于2%的蛋白质对,对应于至少四个生物重复内的FC-A值,我们建议将其作为最小值。如果忠实地遵循程序,可以在3周(瞬时表达)或5周(稳定表达)内在拟南芥中进行酶-酶相互作用的实验评估。©2019作者。基本方案1:基因克隆到目的载体替代方案:In-Fusion或Gibson基因克隆方案基本方案2:将诱饵转化为植物细胞培养或植物叶片基本方案3:蛋白质复合物的亲和纯化基本方案4:头上胰蛋白酶/LysC消化和C18柱肽脱盐和浓缩基本方案5:数据分析和质量控制
本文章由计算机程序翻译,如有差异,请以英文原文为准。
来源期刊
期刊介绍:
Sound and reproducible laboratory methods are the foundation of scientific discovery. Yet nuances that are critical for an experiment''s success are not captured in the primary literature but exist only as part of a lab''s oral tradition. Current Protocols in Plant Biology provides reproducible step-by-step instructions for protocols relevant to plant research. Furthermore, Current Protocols content is thoughtfully organized by topic for optimal usage and to maximize contextual knowledge. Quarterly issues allow Current Protocols in Plant Biology to constantly evolve to keep pace with the newest discoveries and developments. Current Protocols in Plant Biology is the comprehensive source for protocols in the multidisciplinary field of plant biology, providing an extensive range of protocols from basic to cutting edge. Coverage includes: Extraction and analysis of DNA, RNA, proteins Chromosome analysis Transcriptional analysis Protein expression Metabolites Plant enzymology Epigenetics Plant genetic transformation Mutagenesis Arabidopsis, Maize, Poplar, Rice, and Soybean, and more.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


