Integrating gene expression biomarker predictions into networks of adverse outcome pathways
Abstract
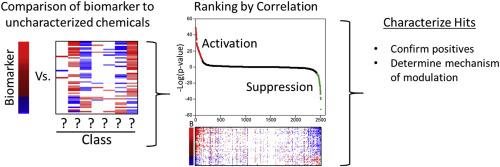
Microarray profiling in the context of toxicity testing in animals has been used for years to identify mechanisms of toxicity, derive points of departure using dose–response modeling, and determine human relevance. High-throughput transcriptomic technologies are increasingly being used to screen environmental chemicals in vitro to identify molecular targets and provide mechanistic context for regulatory testing. This review will discuss the use of gene expression biomarkers to make predictions of activity of molecular targets of chemicals that can be linked to adverse outcomes in a number of cellular and tissue contexts. Gene expression biomarkers are built using global gene expression comparisons from cells or tissues exposed to chemicals with known effects on the factor of interest. Incorporating profiles in which the expression of the factor is altered (e.g. in gene-null mice) facilitates the identification of predictive genes. As an example of their use, biomarkers that predict molecular initiating events and key events in liver cancer adverse outcome pathways have been shown to accurately identify chemical–dose combinations in short-term studies that lead to liver cancer in 2-year bioassays. In the near future, batteries of biomarkers that predict modulation of important targets of environmental chemicals could be used to interpret high-throughput transcriptomic screening data.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


