Single-cell meta-analysis of inflammatory bowel disease with scIBD
IF 18.3
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
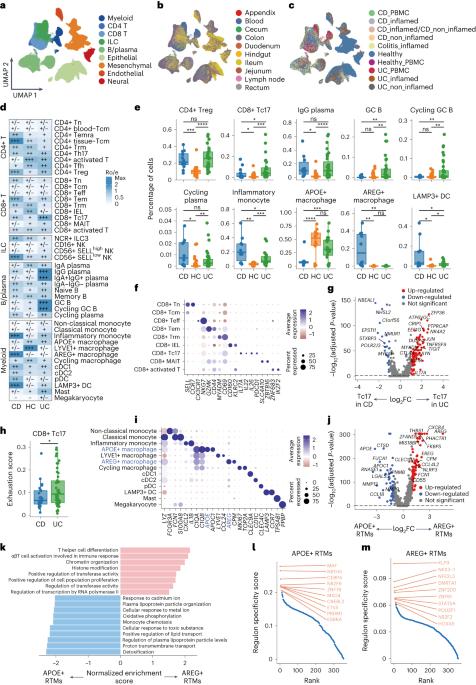
Understanding the heterogeneous intestinal microenvironment is critical to uncover the pathogenesis of inflammatory bowel disease (IBD). Recent advances in single-cell RNA sequencing (scRNA-seq) have identified certain cell types and genes that could contribute to IBD; however, a comprehensively integrated analysis of these scRNA-seq datasets is not yet available. Here we introduce scIBD, a platform for single-cell meta-analysis of IBD with interactive and visualization features, which combines highly curated single-cell datasets in a uniform workflow, enabling identifying rare or less-characterized cell types in IBD and dissecting the commonalities, as well as the differences between ulcerative colitis and Crohn’s disease. scIBD also incorporates multifunctional information—including regulon activity, GWAS-implicated risk genes and genes targeted by therapeutics—to infer clinically relevant cell-type specificity. Collectively, scIBD is a user-friendly web-based platform for the community to analyze the transcriptome features and gene regulatory networks associated with the pathogenesis and treatment of IBD at single-cell resolution. A platform for single-cell meta-analysis of inflammatory bowel disease, named scIBD, enables identification of rare or less-characterized cell types and the dissection of the commonalities and differences between ulcerative colitis and Crohn’s disease.
炎症性肠病伴scIBD的单细胞meta分析
了解异质性肠道微环境对于揭示炎症性肠病(IBD)的发病机制至关重要。单细胞 RNA 测序(scRNA-seq)的最新进展已经确定了可能导致 IBD 的某些细胞类型和基因;然而,目前还没有对这些 scRNA-seq 数据集进行全面综合的分析。在这里,我们介绍了具有交互式和可视化功能的 IBD 单细胞荟萃分析平台 scIBD,该平台在统一的工作流程中结合了高度策划的单细胞数据集,能够识别 IBD 中罕见或特征较少的细胞类型,剖析溃疡性结肠炎和克罗恩病之间的共性和差异。总之,scIBD 是一个用户友好型网络平台,可供社区以单细胞分辨率分析与 IBD 发病和治疗相关的转录组特征和基因调控网络。被命名为 scIBD 的炎症性肠病单细胞荟萃分析平台能够识别罕见或特征较少的细胞类型,并剖析溃疡性结肠炎和克罗恩病之间的共性和差异。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


