Capturing long‐tailed individual tree diversity using an airborne imaging and a multi‐temporal hierarchical model
IF 3.9
2区 环境科学与生态学
Q1 ECOLOGY
引用次数: 2
Abstract
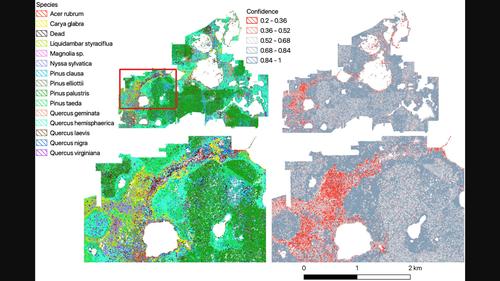
Measuring forest biodiversity using terrestrial surveys is expensive and can only capture common species abundance in large heterogeneous landscapes. In contrast, combining airborne imagery with computer vision can generate individual tree data at the scales of hundreds of thousands of trees. To train computer vision models, ground‐based species labels are combined with airborne reflectance data. Due to the difficulty of finding rare species in a large landscape, many classification models only include the most abundant species, leading to biased predictions at broad scales. For example, if only common species are used to train the model, this assumes that these samples are representative across the entire landscape. Extending classification models to include rare species requires targeted data collection and algorithmic improvements to overcome large data imbalances between dominant and rare taxa. We use a targeted sampling workflow to the Ordway Swisher Biological Station within the US National Ecological Observatory Network (NEON), where traditional forestry plots had identified six canopy tree species with more than 10 individuals at the site. Combining iterative model development with rare species sampling, we extend a training dataset to include 14 species. Using a multi‐temporal hierarchical model, we demonstrate the ability to include species predicted at <1% frequency in landscape without losing performance on the dominant species. The final model has over 75% accuracy for 14 species with improved rare species classification compared to 61% accuracy of a baseline deep learning model. After filtering out dead trees, we generate landscape species maps of individual crowns for over 670 000 individual trees. We find distinct patches of forest composed of rarer species at the full‐site scale, highlighting the importance of capturing species diversity in training data. We estimate the relative abundance of 14 species within the landscape and provide three measures of uncertainty to generate a range of counts for each species. For example, we estimate that the dominant species, Pinus palustris accounts for c. 28% of predicted stems, with models predicting a range of counts between 160 000 and 210 000 individuals. These maps provide the first estimates of canopy tree diversity within a NEON site to include rare species and provide a blueprint for capturing tree diversity using airborne computer vision at broad scales.
使用航空成像和多时相分层模型捕捉长尾个体树木多样性
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Remote Sensing in Ecology and Conservation
Earth and Planetary Sciences-Computers in Earth Sciences
CiteScore
9.80
自引率
5.50%
发文量
69
审稿时长
18 weeks
期刊介绍:
emote Sensing in Ecology and Conservation provides a forum for rapid, peer-reviewed publication of novel, multidisciplinary research at the interface between remote sensing science and ecology and conservation. The journal prioritizes findings that advance the scientific basis of ecology and conservation, promoting the development of remote-sensing based methods relevant to the management of land use and biological systems at all levels, from populations and species to ecosystems and biomes. The journal defines remote sensing in its broadest sense, including data acquisition by hand-held and fixed ground-based sensors, such as camera traps and acoustic recorders, and sensors on airplanes and satellites. The intended journal’s audience includes ecologists, conservation scientists, policy makers, managers of terrestrial and aquatic systems, remote sensing scientists, and students.
Remote Sensing in Ecology and Conservation is a fully open access journal from Wiley and the Zoological Society of London. Remote sensing has enormous potential as to provide information on the state of, and pressures on, biological diversity and ecosystem services, at multiple spatial and temporal scales. This new publication provides a forum for multidisciplinary research in remote sensing science, ecological research and conservation science.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


