Novel Gene Signatures for Prostate Cancer Detection: Network Centrality-based Screening with Experimental Validation
IF 2.9
3区 生物学
Q3 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Prostate cancer (PCa) is a kind of malignant tumor with high incidence among males worldwide. The identification of novel biomarker signatures is, therefore of clinical significance for PCa precision medicine. It has been acknowledged that the breaking of stability and vulnerability in biological network provides important clues for cancer biomarker discovery. In this study, a bioinformatics model by characterizing the centrality of nodes in PCa-specific protein-protein interaction (PPI) network was proposed and applied to identify novel gene signatures for PCa detection. Compared with traditional methods, this model integrated degree, closeness and betweenness centrality as the criterion for Hub gene prioritization. The identified biomarkers were validated based on receiver-operating characteristic evaluation, qRT-PCR experimental analysis and literature-guided functional survey. Four genes, i.e., MYOF, RBFOX3, OCLN, and CDKN1C, were screened with average AUC ranging from 0.79 to 0.87 in the predicted and validated datasets for PCa diagnosis. Among them, MYOF, RBFOX3, and CDKN1C were observed to be down-regulated whereas OCLN was over-expressed in PCa groups. The in vitro qRT-PCR experiment using cell line samples convinced the potential of identified genes as novel biomarkers for PCa detection. Biological process and pathway enrichment analysis suggested the underlying role of identified biomarkers in mediating PCa-related genes and pathways including TGF-β, Hippo, MAPK signaling during PCa occurrence and progression. Novel gene signatures were screened as candidate biomarkers for PCa detection based on topological characterization of PCa-specific PPI network. More clinical validation using human samples will be performed in future work.
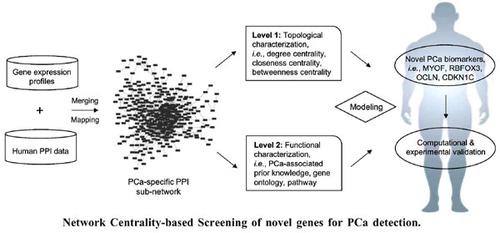
前列腺癌检测的新基因特征:基于网络中心性的筛选与实验验证
癌症(PCa)是一种世界范围内男性恶性肿瘤,发病率较高。因此,识别新的生物标志物特征对前列腺癌精准医学具有临床意义。人们已经认识到,生物网络稳定性和脆弱性的打破为癌症生物标志物的发现提供了重要线索。在本研究中,通过表征PCa特异性蛋白质-蛋白质相互作用(PPI)网络中节点的中心性,提出了一个生物信息学模型,并将其应用于识别PCa检测的新基因特征。与传统方法相比,该模型综合了程度、贴近度和介数中心性作为Hub基因优先的标准。基于受试者操作特征评估、qRT-PCR实验分析和文献引导的功能调查,对已鉴定的生物标志物进行了验证。在预测和验证的PCa诊断数据集中,筛选了四个基因,即MYOF、RBFOX3、OCLN和CDKN1C,平均AUC范围为0.79至0.87。其中,MYOF、RBFOX3和CDKN1C被观察到下调,而OCLN在PCa组中过度表达。使用细胞系样本的体外qRT-PCR实验证实了已鉴定基因作为PCa检测新生物标志物的潜力。生物学过程和通路富集分析表明,已鉴定的生物标志物在PCa发生和发展过程中介导PCa相关基因和通路(包括TGF-β、Hippo、MAPK信号传导)的潜在作用。基于PCa特异性PPI网络的拓扑特征,筛选新的基因特征作为PCa检测的候选生物标志物。在未来的工作中,将使用人体样本进行更多的临床验证。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current Bioinformatics
生物-生化研究方法
CiteScore
6.60
自引率
2.50%
发文量
77
审稿时长
>12 weeks
期刊介绍:
Current Bioinformatics aims to publish all the latest and outstanding developments in bioinformatics. Each issue contains a series of timely, in-depth/mini-reviews, research papers and guest edited thematic issues written by leaders in the field, covering a wide range of the integration of biology with computer and information science.
The journal focuses on advances in computational molecular/structural biology, encompassing areas such as computing in biomedicine and genomics, computational proteomics and systems biology, and metabolic pathway engineering. Developments in these fields have direct implications on key issues related to health care, medicine, genetic disorders, development of agricultural products, renewable energy, environmental protection, etc.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


