Pharmacogenomics of Clozapine-induced agranulocytosis: a systematic review and meta-analysis
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 6
Abstract
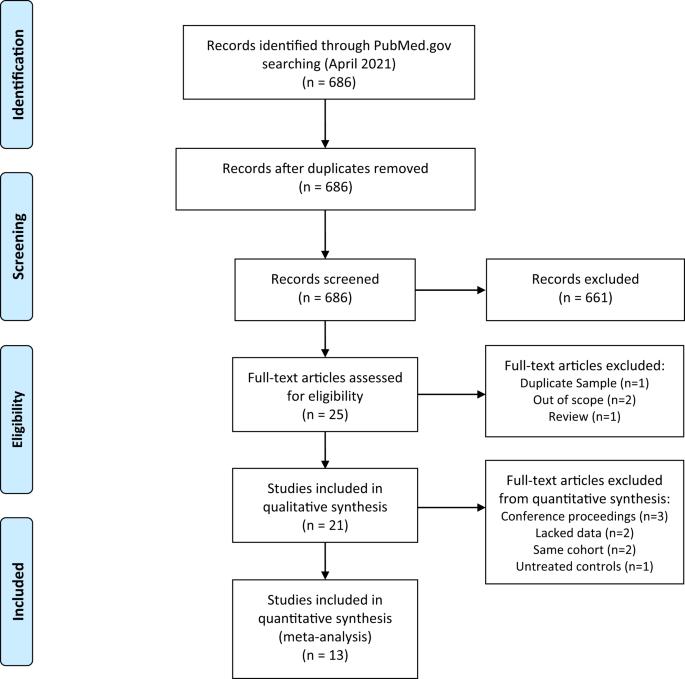
Although clozapine is the most effective pharmacotherapy for treatment-resistant schizophrenia, it is under-utilized, and initiation is often delayed. One reason is the occurrence of a potentially fatal adverse reaction, clozapine-induced agranulocytosis (CIA). Identifying genetic variations contributing to CIA would help predict patient risk of developing CIA and personalize treatment. Here, we (1) review existing pharmacogenomic studies of CIA, and (2) conduct meta-analyses to identify targets for clinical implementation. A systematic literature search identified studies that included individuals receiving clozapine who developed CIA and controls who did not. Results showed that individuals carrying the HLA-DRB1*04:02 allele had nearly sixfold (95% CI 2.20–15.80, pcorrected = 0.03) higher odds of CIA with a negative predictive value of 99.3%. Previously unreplicated alleles, TNFb5, HLA-B*59:01, TNFb4, and TNFd3 showed significant associations with CIA after multiple-testing corrections. Our findings suggest that a predictive HLA-DRB1*04:02-based pharmacogenomic test may be promising for clinical implementation but requires further investigation.
氯氮平诱导的粒细胞缺乏症的药物基因组学:系统回顾和荟萃分析
尽管氯氮平是治疗耐药性精神分裂症最有效的药物疗法,但它的使用率却很低,而且常常被推迟使用。其中一个原因是出现了一种可能致命的不良反应--氯氮平诱发的粒细胞减少症(CIA)。确定导致 CIA 的基因变异有助于预测患者罹患 CIA 的风险并进行个性化治疗。在此,我们(1)回顾了现有的 CIA 药物基因组学研究,(2)进行了荟萃分析,以确定临床实施的目标。通过系统性文献检索,我们找到了包括接受氯氮平治疗但出现 CIA 的患者和未出现 CIA 的对照组的研究。结果显示,携带HLA-DRB1*04:02等位基因的个体发生CIA的几率高出近6倍(95% CI 2.20-15.80, pcorrected = 0.03),阴性预测值为99.3%。之前未被复制的等位基因 TNFb5、HLA-B*59:01、TNFb4 和 TNFd3 在多重检验校正后显示与 CIA 有显著关联。我们的研究结果表明,基于 HLA-DRB1*04:02 的预测性药物基因组学检测可能有望在临床上应用,但还需要进一步研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


