Fine mapping of the Cepaea nemoralis shell colour and mid-banded loci using a high-density linkage map
IF 3.1
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
Molluscs are a highly speciose phylum that exhibits an astonishing array of colours and patterns, yet relatively little progress has been made in identifying the underlying genes that determine phenotypic variation. One prominent example is the land snail Cepaea nemoralis for which classical genetic studies have shown that around nine loci, several physically linked and inherited together as a ‘supergene’, control the shell colour and banding polymorphism. As a first step towards identifying the genes involved, we used whole-genome resequencing of individuals from a laboratory cross to construct a high-density linkage map, and then trait mapping to identify 95% confidence intervals for the chromosomal region that contains the supergene, specifically the colour locus (C), and the unlinked mid-banded locus (U). The linkage map is made up of 215,593 markers, ordered into 22 linkage groups, with one large group making up ~27% of the genome. The C locus was mapped to a ~1.3 cM region on linkage group 11, and the U locus was mapped to a ~0.7 cM region on linkage group 15. The linkage map will serve as an important resource for further evolutionary and population genomic studies of C. nemoralis and related species, as well as the identification of candidate genes within the supergene and for the mid-banding phenotype.
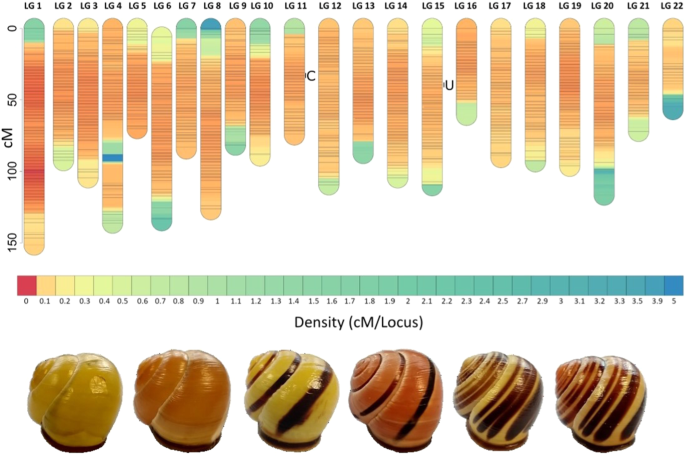
利用高密度连锁图谱精细定位内莫拉氏Cepaea外壳颜色和中带基因座。
软体动物是一个高度物种化的门,具有惊人的颜色和模式,但在确定决定表型变异的潜在基因方面进展相对较小。一个突出的例子是陆地蜗牛Cepaea nemoralis,其经典遗传学研究表明,大约有九个基因座控制着外壳颜色和条带多态性,其中几个基因座作为“表基因”物理连接并遗传在一起。作为识别相关基因的第一步,我们使用实验室杂交个体的全基因组重新测序来构建高密度连锁图谱,然后使用性状图谱来识别包含超基因的染色体区域的95%置信区间,特别是颜色基因座(C)和未连接的中带基因座(U)。连锁图谱由215593个标记组成,分为22个连锁组,其中一个大组占基因组的27%。C基因座被定位到a~1.3 连锁群11上的cM区,U位点被定位到a~0.7 连锁群15上的cM区。该连锁图将作为一个重要的资源,用于进一步研究奈莫拉氏菌和相关物种的进化和种群基因组,以及鉴定表基因内的候选基因和中带表型。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


