Development of a Multiepitope Vaccine Against SARS-CoV-2: Immunoinformatics Study.
JMIR bioinformatics and biotechnology
Pub Date : 2022-07-19
eCollection Date: 2022-01-01
DOI:10.2196/36100
引用次数: 4
Abstract
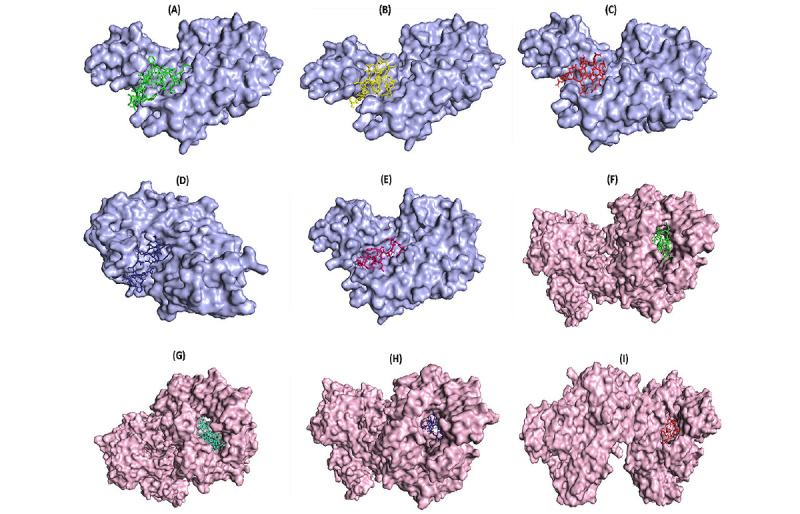
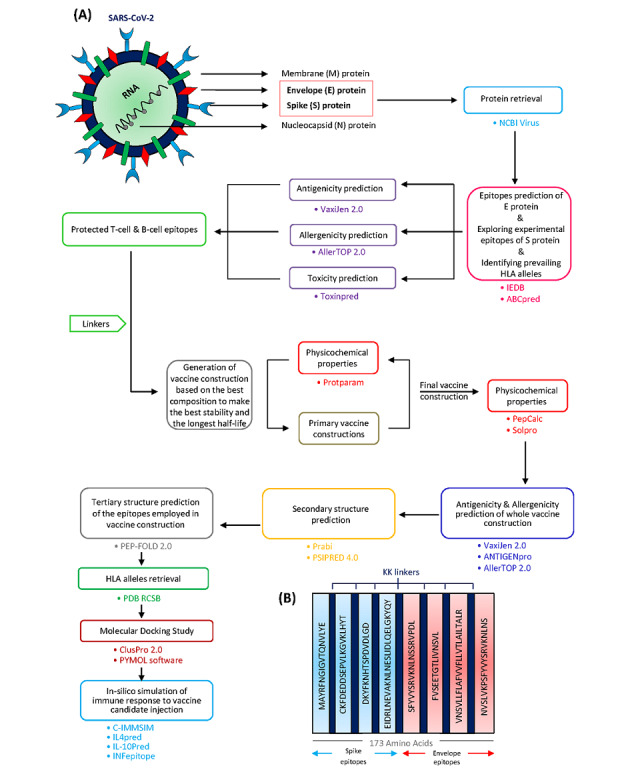
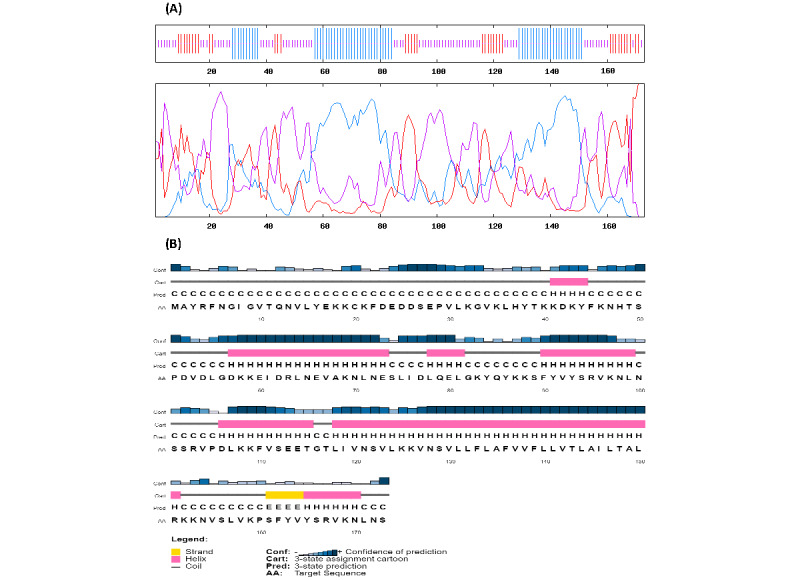
Background Since the first appearance of SARS-CoV-2 in China in December 2019, the world witnessed the emergence of the SARS-CoV-2 outbreak. Due to the high transmissibility rate of the virus, there is an urgent need to design and develop vaccines against SARS-CoV-2 to prevent more cases affected by the virus. Objective A computational approach is proposed for vaccine design against the SARS-CoV-2 spike (S) protein, as the key target for neutralizing antibodies, and envelope (E) protein, which contains a conserved sequence feature. Methods We used previously reported epitopes of S protein detected experimentally and further identified a collection of predicted B-cell and major histocompatibility (MHC) class II–restricted T-cell epitopes derived from E proteins with an identical match to SARS-CoV-2 E protein. Results The in silico design of our candidate vaccine against the S and E proteins of SARS-CoV-2 demonstrated a high affinity to MHC class II molecules and effective results in immune response simulations. Conclusions Based on the results of this study, the multiepitope vaccine designed against the S and E proteins of SARS-CoV-2 may be considered as a new, safe, and efficient approach to combatting the COVID-19 pandemic.
SARS-CoV-2多表位疫苗的研制:免疫信息学研究
背景:自2019年12月中国首次出现SARS-CoV-2以来,世界见证了SARS-CoV-2疫情的出现。由于该病毒的高传播率,迫切需要设计和开发针对SARS-CoV-2的疫苗,以防止更多的病例感染该病毒。目的:提出了一种针对SARS-CoV-2刺突蛋白(S)和包膜蛋白(E)的疫苗设计计算方法,S蛋白是中和抗体的关键靶点,E蛋白具有保守序列特征。方法:利用先前报道的实验检测到的S蛋白表位,进一步鉴定了来自E蛋白的预测b细胞和主要组织相容性(MHC) ii类限制性t细胞表位,这些表位与SARS-CoV-2 E蛋白完全匹配。结果:我们设计的SARS-CoV-2 S和E蛋白候选疫苗与MHC II类分子具有高亲和力,在免疫反应模拟中取得了有效的结果。结论:基于本研究结果,针对SARS-CoV-2的S和E蛋白设计的多表位疫苗可能被认为是一种新的、安全的、有效的对抗COVID-19大流行的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


