Whole-exome sequencing reveals rare genetic variations in ovarian granulosa cell tumor.
IF 3.4
4区 医学
Q2 MEDICINE, RESEARCH & EXPERIMENTAL
引用次数: 2
Abstract
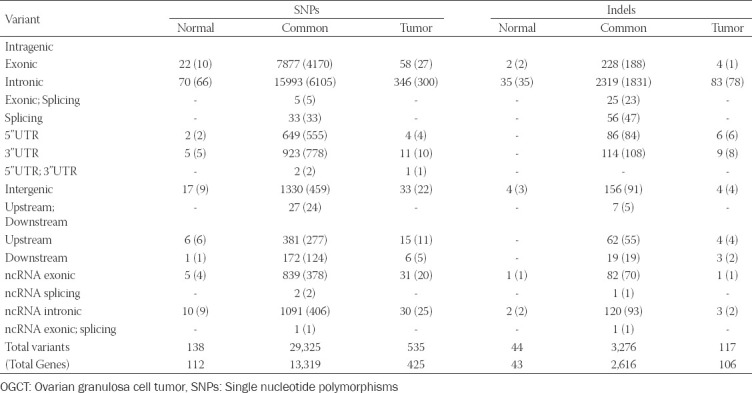
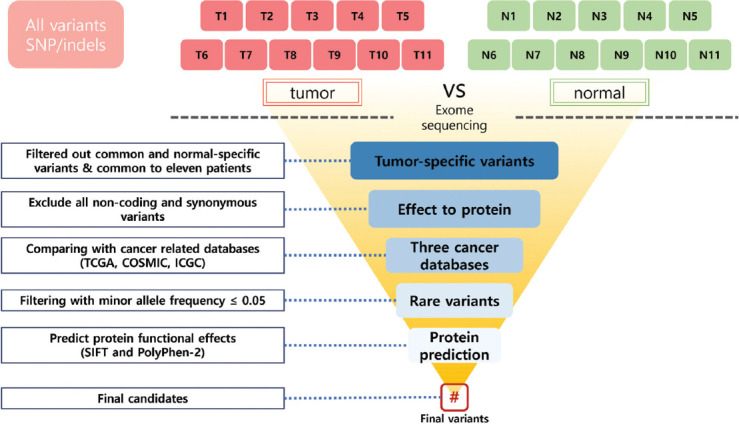
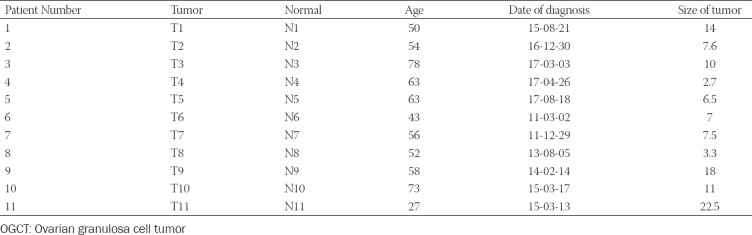
Ovarian granulosa cell tumor (OGCT) is a rare ovarian tumor that accounts for about 2-5% of all ovarian tumors. Despite the low grade of ovarian tumors, high and late recurrences are common in OGCT patients. Even though this tumor usually occurs in adult women with high estrogen levels, the cause of OGCT is still unknown. To screen genetic variants associated with OGCT, we collected normal and matched-tumor formalin-fixed paraffin-embedded from 11 OGCT patients and performed whole-exome sequencing using Illumina NovaSeq 6000. A total of 1,067,219 single nucleotide polymorphisms (SNPs) and 162,155 insertions/deletions (indels) were identified from 11 pairs of samples. Of these, we identified 44 tumor-specific SNPs in 22 genes and four tumor-specific indels in one gene that were common to 11 patients. We used three cancer databases (TCGA, COSMIC, and ICGC) to investigate genes associated with ovarian cancers. Nine genes (SEC22B, FEZ2, ANKRD36B, GYPA, MUC3A, PRSS3, NUTM2A, OR8U1, and KRTAP10-6) associated with ovarian cancers were found in all three databases. In addition, we identified seven rare variants with MAF ≤ 0.05 in two genes (PRSS3 and MUC3A). Of seven rare variants, five variants in MUC3A are potentially pathogenic. Furthermore, we conducted gene enrichment analysis of tumor-specific 417 genes in SNPs and 106 genes in indels using cytoscape and metascape. In GO analysis, these genes were highly enriched in “selective autophagy,” and “regulation of anoikis.” Taken together, we suggest that MUC3A is implicated in OGCT development, and MUC3A could be used as a potential biomarker for OGCT diagnosis.
全外显子组测序揭示卵巢颗粒细胞瘤罕见的遗传变异。
卵巢颗粒细胞瘤(OGCT)是一种罕见的卵巢肿瘤,约占卵巢肿瘤的2-5%。尽管卵巢肿瘤的分级低,但高复发和晚期复发在OGCT患者中很常见。尽管这种肿瘤通常发生在雌激素水平高的成年女性,但导致OGCT的原因尚不清楚。为了筛选与OGCT相关的遗传变异,我们从11名OGCT患者中收集了正常和匹配肿瘤的福尔马林固定石蜡包埋(FFPE),并使用Illumina NovaSeq 6000进行了全外展子组测序(WES)。从11对样本中共鉴定出1,067,219个单核苷酸多态性(snp)和162,155个插入/缺失(indels)。其中,我们鉴定了22个基因中的44个肿瘤特异性snp和11个患者共有的一个基因中的4个肿瘤特异性indel。我们使用三个癌症数据库(TCGA、COSMIC和ICGC)来研究与卵巢癌相关的基因。在这三个数据库中均发现了9个与卵巢癌相关的基因(SEC22B、FEZ2、ANKRD36B、GYPA、MUC3A、PRSS3、NUTM2A、OR8U1和KRTAP10-6)。此外,我们在两个基因(PRSS3和MUC3A)中发现了7个MAF≤0.05的罕见变异。在7个罕见变异中,MUC3A的5个变异具有潜在致病性。此外,我们利用细胞景观和元景观对SNPs中的417个肿瘤特异性基因和indels中的106个基因进行了基因富集分析。在氧化石墨烯分析中,这些基因在“选择性自噬”和“anoikis调控”中高度富集。综上所述,我们认为MUC3A与OGCT的发展有关,MUC3A可以作为OGCT诊断的潜在生物标志物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Bosnian journal of basic medical sciences
医学-医学:研究与实验
CiteScore
7.40
自引率
5.90%
发文量
98
审稿时长
35 days
期刊介绍:
The Bosnian Journal of Basic Medical Sciences (BJBMS) is an international, English-language, peer reviewed journal, publishing original articles from different disciplines of basic medical sciences. BJBMS welcomes original research and comprehensive reviews as well as short research communications in the field of biochemistry, genetics, immunology, microbiology, pathology, pharmacology, pharmaceutical sciences and physiology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


