Analysis of primitive genetic interactions for the design of a genetic signal differentiator.
IF 2.6
Q2 BIOCHEMICAL RESEARCH METHODS
Synthetic biology (Oxford, England)
Pub Date : 2019-06-27
eCollection Date: 2019-01-01
DOI:10.1093/synbio/ysz015
引用次数: 6
Abstract
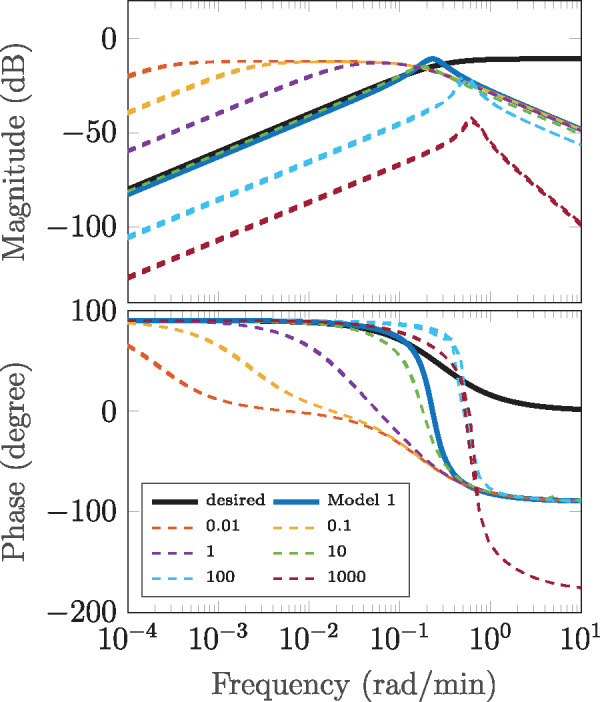
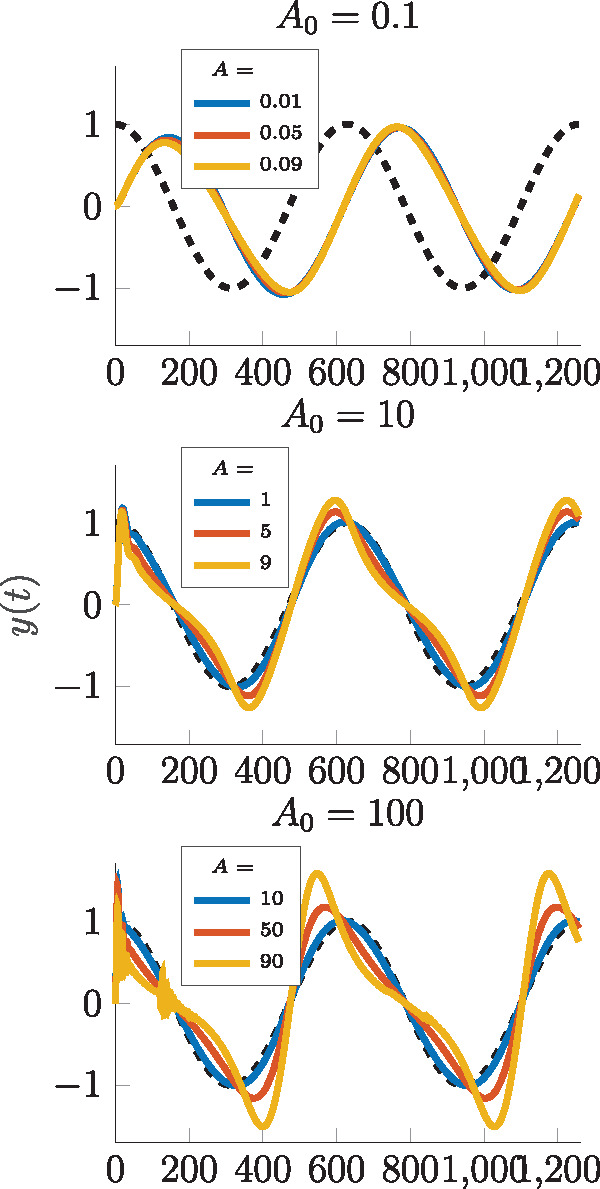
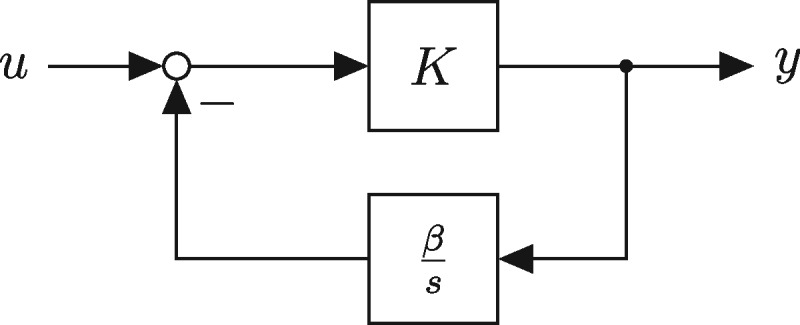
Abstract We study the dynamic and static input–output behavior of several primitive genetic interactions and their effect on the performance of a genetic signal differentiator. In a simplified design, several requirements for the linearity and time-scales of processes like transcription, translation and competitive promoter binding were introduced. By experimentally probing simple genetic constructs in a cell-free experimental environment and fitting semi-mechanistic models to these data, we show that some of these requirements can be verified, while others are only met with reservations in certain operational regimes. Analyzing the linearized model of the resulting genetic network, we conclude that it approximates a differentiator with relative degree one. Taking also the discovered nonlinearities into account and using a describing function approach, we further determine the particular frequency and amplitude ranges where the genetic differentiator can be expected to behave as such.
遗传信号微分器设计中的原始遗传相互作用分析。
研究了几种原始遗传相互作用的动态和静态输入输出行为及其对遗传信号微分器性能的影响。在一个简化的设计中,介绍了对转录、翻译和竞争性启动子结合等过程的线性和时间尺度的几个要求。通过在无细胞实验环境中实验探测简单的遗传结构,并将半机械模型拟合到这些数据中,我们表明其中一些要求可以得到验证,而其他要求仅在某些操作制度下得到保留。分析所得遗传网络的线性化模型,我们得出结论,它近似于一个相对度为1的微分器。考虑到发现的非线性并使用描述函数方法,我们进一步确定遗传微分器可以预期表现的特定频率和幅度范围。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


