Hannah Döpper, Julia Menges, Morgane Bozet, Alexandra Brenzel, Dietmar Lohmann, Laura Steenpass, Deniz Kanber
下载PDF
{"title":"Differentiation Protocol for 3D Retinal Organoids, Immunostaining and Signal Quantitation","authors":"Hannah Döpper, Julia Menges, Morgane Bozet, Alexandra Brenzel, Dietmar Lohmann, Laura Steenpass, Deniz Kanber","doi":"10.1002/cpsc.120","DOIUrl":null,"url":null,"abstract":"<p>Structures resembling whole organs, called organoids, are generated using pluripotent stem cells and 3D culturing methods. This relies on the ability of cells to self-reorganize after dissociation. In combination with certain supplemented factors, differentiation can be directed toward the formation of several organ-like structures. Here, a protocol for the generation of retinal organoids containing all seven retinal cell types is described. This protocol does not depend on Matrigel, and by keeping the organoids single and independent at all times, fusion is prevented and monitoring of differentiation is improved. Comprehensive phenotypic characterization of the in vitro–generated retinal organoids is achieved by the protocol for immunostaining outlined here. By comparing different stages of retinal organoids, the decrease and increase of certain cell populations can be determined. In order to be able to detect even small differences, it is necessary to quantify the immunofluorescent signals, for which we have provided a detailed protocol describing signal quantitation using the image-processing program Fiji. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: Differentiation protocol for 3D retinal organoids</p><p><b>Basic Protocol 2</b>: Immunostaining protocol for cryosections of retinal organoids</p><p><b>Support Protocol</b>: Embedding and sectioning protocol for 3D retinal organoids</p><p><b>Basic Protocol 3</b>: Quantitation protocol using Fiji</p>","PeriodicalId":53703,"journal":{"name":"Current Protocols in Stem Cell Biology","volume":"55 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-09-21","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpsc.120","citationCount":"8","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current Protocols in Stem Cell Biology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpsc.120","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q2","JCRName":"Biochemistry, Genetics and Molecular Biology","Score":null,"Total":0}
引用次数: 8
引用
批量引用
Abstract
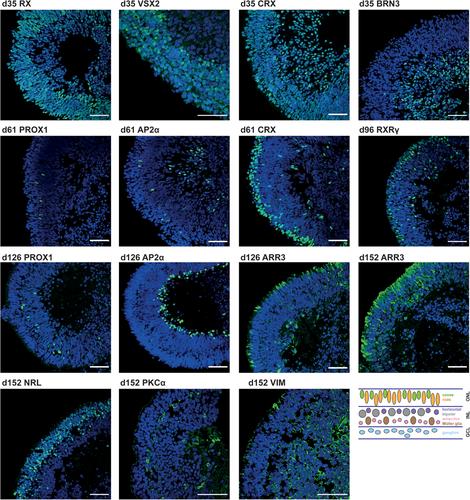
Structures resembling whole organs, called organoids, are generated using pluripotent stem cells and 3D culturing methods. This relies on the ability of cells to self-reorganize after dissociation. In combination with certain supplemented factors, differentiation can be directed toward the formation of several organ-like structures. Here, a protocol for the generation of retinal organoids containing all seven retinal cell types is described. This protocol does not depend on Matrigel, and by keeping the organoids single and independent at all times, fusion is prevented and monitoring of differentiation is improved. Comprehensive phenotypic characterization of the in vitro–generated retinal organoids is achieved by the protocol for immunostaining outlined here. By comparing different stages of retinal organoids, the decrease and increase of certain cell populations can be determined. In order to be able to detect even small differences, it is necessary to quantify the immunofluorescent signals, for which we have provided a detailed protocol describing signal quantitation using the image-processing program Fiji. © 2020 The Authors.
Basic Protocol 1 : Differentiation protocol for 3D retinal organoids
Basic Protocol 2 : Immunostaining protocol for cryosections of retinal organoids
Support Protocol : Embedding and sectioning protocol for 3D retinal organoids
Basic Protocol 3 : Quantitation protocol using Fiji
三维视网膜类器官分化方案,免疫染色和信号定量
类似于整个器官的结构,称为类器官,是用多能干细胞和3D培养方法产生的。这依赖于细胞在分离后自我重组的能力。在某些补充因子的作用下,分化可指向几种器官样结构的形成。在这里,一个方案的产生视网膜类器官包含所有七种视网膜细胞类型被描述。该方案不依赖于Matrigel,并且通过始终保持类器官的单一和独立,可以防止融合并改善对分化的监测。体外生成的视网膜类器官的综合表型表征是通过这里概述的免疫染色方案实现的。通过比较不同阶段的视网膜类器官,可以确定某些细胞群的减少和增加。为了能够检测到甚至很小的差异,有必要对免疫荧光信号进行量化,为此,我们提供了使用图像处理程序Fiji描述信号量化的详细协议。©2020作者。基本方案1:3D视网膜类器官的分化方案基本方案2:视网膜类器官冷冻切片的免疫染色方案支持方案:3D视网膜类器官的嵌入和切片方案基本方案3:使用Fiji的定量方案
本文章由计算机程序翻译,如有差异,请以英文原文为准。

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


