Marie A. Brunet, Amina M. Lekehal, Xavier Roucou
下载PDF
{"title":"How to Illuminate the Dark Proteome Using the Multi-omic OpenProt Resource","authors":"Marie A. Brunet, Amina M. Lekehal, Xavier Roucou","doi":"10.1002/cpbi.103","DOIUrl":null,"url":null,"abstract":"<p>Ten of thousands of open reading frames (ORFs) are hidden within genomes. These alternative ORFs, or small ORFs, have eluded annotations because they are either small or within unsuspected locations. They are found in untranslated regions or overlap a known coding sequence in messenger RNA and anywhere in a “non-coding” RNA. Serendipitous discoveries have highlighted these ORFs’ importance in biological functions and pathways. With their discovery came the need for deeper ORF annotation and large-scale mining of public repositories to gather supporting experimental evidence. OpenProt, accessible at https://openprot.org/, is the first proteogenomic resource enforcing a polycistronic model of annotation across an exhaustive transcriptome for 10 species. Moreover, OpenProt reports experimental evidence cumulated across a re-analysis of 114 mass spectrometry and 87 ribosome profiling datasets. The multi-omics OpenProt resource also includes the identification of predicted functional domains and evaluation of conservation for all predicted ORFs. The OpenProt web server provides two query interfaces and one genome browser. The query interfaces allow for exploration of the coding potential of genes or transcripts of interest as well as custom downloads of all information contained in OpenProt. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: Using the Search interface</p><p><b>Basic Protocol 2</b>: Using the Downloads interface</p>","PeriodicalId":10958,"journal":{"name":"Current protocols in bioinformatics","volume":"71 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-08-11","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpbi.103","citationCount":"3","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in bioinformatics","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpbi.103","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Biochemistry, Genetics and Molecular Biology","Score":null,"Total":0}
引用次数: 3
引用
批量引用
Abstract
Ten of thousands of open reading frames (ORFs) are hidden within genomes. These alternative ORFs, or small ORFs, have eluded annotations because they are either small or within unsuspected locations. They are found in untranslated regions or overlap a known coding sequence in messenger RNA and anywhere in a “non-coding” RNA. Serendipitous discoveries have highlighted these ORFs’ importance in biological functions and pathways. With their discovery came the need for deeper ORF annotation and large-scale mining of public repositories to gather supporting experimental evidence. OpenProt, accessible at https://openprot.org/, is the first proteogenomic resource enforcing a polycistronic model of annotation across an exhaustive transcriptome for 10 species. Moreover, OpenProt reports experimental evidence cumulated across a re-analysis of 114 mass spectrometry and 87 ribosome profiling datasets. The multi-omics OpenProt resource also includes the identification of predicted functional domains and evaluation of conservation for all predicted ORFs. The OpenProt web server provides two query interfaces and one genome browser. The query interfaces allow for exploration of the coding potential of genes or transcripts of interest as well as custom downloads of all information contained in OpenProt. © 2020 The Authors.
Basic Protocol 1 : Using the Search interface
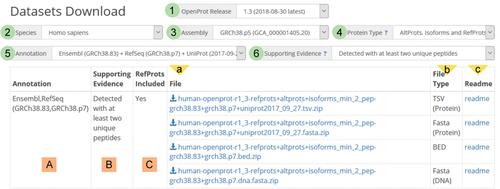
Basic Protocol 2 : Using the Downloads interface
如何利用多基因组开放资源揭示黑暗蛋白质组
基因组中隐藏着成千上万个开放阅读框(orf)。这些备选orf或小型orf都避开了注释,因为它们要么很小,要么位于未知位置。它们存在于信使RNA的非翻译区或与已知编码序列重叠的地方,以及“非编码”RNA的任何地方。偶然的发现突出了这些orf在生物学功能和途径中的重要性。随着他们的发现,需要更深入的ORF注释和对公共存储库的大规模挖掘来收集支持性的实验证据。OpenProt,可访问https://openprot.org/,是第一个蛋白质基因组资源,在10个物种的详尽转录组中执行多顺反子注释模型。此外,OpenProt报告了通过114个质谱分析和87个核糖体分析数据集重新分析积累的实验证据。多组学OpenProt资源还包括预测功能域的鉴定和所有预测orf的保守性评估。OpenProt web服务器提供了两个查询接口和一个基因组浏览器。查询接口允许探索基因的编码潜力或感兴趣的转录本,以及自定义下载OpenProt中包含的所有信息。©2020作者。基本协议1:使用Search接口基本协议2:使用Downloads接口
本文章由计算机程序翻译,如有差异,请以英文原文为准。

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


