Mingzhang Yang, Myra K. Derbyshire, Roxanne A. Yamashita, Aron Marchler-Bauer
下载PDF
{"title":"NCBI's Conserved Domain Database and Tools for Protein Domain Analysis","authors":"Mingzhang Yang, Myra K. Derbyshire, Roxanne A. Yamashita, Aron Marchler-Bauer","doi":"10.1002/cpbi.90","DOIUrl":null,"url":null,"abstract":"<p>The Conserved Domain Database (CDD) is a freely available resource for the annotation of sequences with the locations of conserved protein domain footprints, as well as functional sites and motifs inferred from these footprints. It includes protein domain and protein family models curated in house by CDD staff, as well as imported from a variety of other sources. The latest CDD release (v3.17, April 2019) contains more than 57,000 domain models, of which almost 15,000 were curated by CDD staff. The CDD curation effort increases coverage and provides finer-grained classifications of common and widely distributed protein domain families, for which a wealth of functional and structural data have become available. The CDD maintains both live search capabilities and an archive of pre-computed domain annotations for a selected subset of sequences tracked by the NCBI's Entrez protein database. These can be retrieved or computed for a single sequence using CD-Search or in bulk using Batch CD-Search, or computed via standalone RPS-BLAST plus the rpsbproc software package. The CDD can be accessed via https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. The three protocols listed here describe how to perform a CD-Search (Basic Protocol 1), a Batch CD-Search (Basic Protocol 2), and a Standalone RPS-BLAST and rpsbproc (Basic Protocol 3). © 2019 The Authors.</p><p><b>Basic Protocol 1</b>: CD-search</p><p><b>Basic Protocol 2</b>: Batch CD-search</p><p><b>Basic Protocol 3</b>: Standalone RPS-BLAST and rpsbproc</p>","PeriodicalId":10958,"journal":{"name":"Current protocols in bioinformatics","volume":"69 1","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2019-12-18","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpbi.90","citationCount":"107","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in bioinformatics","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpbi.90","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Biochemistry, Genetics and Molecular Biology","Score":null,"Total":0}
引用次数: 107
引用
批量引用
Abstract
The Conserved Domain Database (CDD) is a freely available resource for the annotation of sequences with the locations of conserved protein domain footprints, as well as functional sites and motifs inferred from these footprints. It includes protein domain and protein family models curated in house by CDD staff, as well as imported from a variety of other sources. The latest CDD release (v3.17, April 2019) contains more than 57,000 domain models, of which almost 15,000 were curated by CDD staff. The CDD curation effort increases coverage and provides finer-grained classifications of common and widely distributed protein domain families, for which a wealth of functional and structural data have become available. The CDD maintains both live search capabilities and an archive of pre-computed domain annotations for a selected subset of sequences tracked by the NCBI's Entrez protein database. These can be retrieved or computed for a single sequence using CD-Search or in bulk using Batch CD-Search, or computed via standalone RPS-BLAST plus the rpsbproc software package. The CDD can be accessed via https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. The three protocols listed here describe how to perform a CD-Search (Basic Protocol 1), a Batch CD-Search (Basic Protocol 2), and a Standalone RPS-BLAST and rpsbproc (Basic Protocol 3). © 2019 The Authors.
Basic Protocol 1 : CD-search
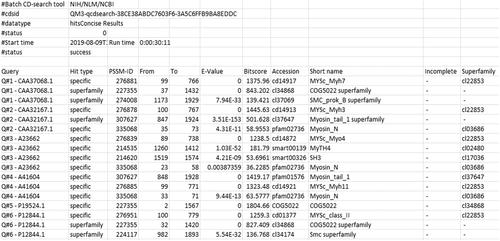
Basic Protocol 2 : Batch CD-search
Basic Protocol 3 : Standalone RPS-BLAST and rpsbproc
NCBI的保守结构域数据库和蛋白质结构域分析工具
保守结构域数据库(CDD)是一个免费的资源,用于标注具有保守蛋白结构域足迹位置的序列,以及从这些足迹推断出的功能位点和基序。它包括由CDD工作人员在内部策划的蛋白质结构域和蛋白质家族模型,以及从各种其他来源导入的模型。最新的CDD版本(v3.17, 2019年4月)包含超过57,000个域模型,其中近15,000个由CDD工作人员策划。CDD管理工作增加了覆盖范围,并提供了对常见和广泛分布的蛋白质结构域家族的更细粒度的分类,为此提供了丰富的功能和结构数据。CDD维护实时搜索功能和NCBI的Entrez蛋白质数据库跟踪的选定序列子集的预计算域注释存档。这些数据可以使用CD-Search检索或计算单个序列,也可以使用Batch CD-Search批量检索或计算,或者通过独立的RPS-BLAST加上rpsbproc软件包进行计算。CDD可以通过https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml访问。这里列出的三个协议描述了如何执行cd搜索(基本协议1),批量cd搜索(基本协议2)以及独立RPS-BLAST和rpsbproc(基本协议3)。©2019作者。基本协议1:cd -search基本协议2:批量cd -search基本协议3:独立的RPS-BLAST和rpsbproc
本文章由计算机程序翻译,如有差异,请以英文原文为准。

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


